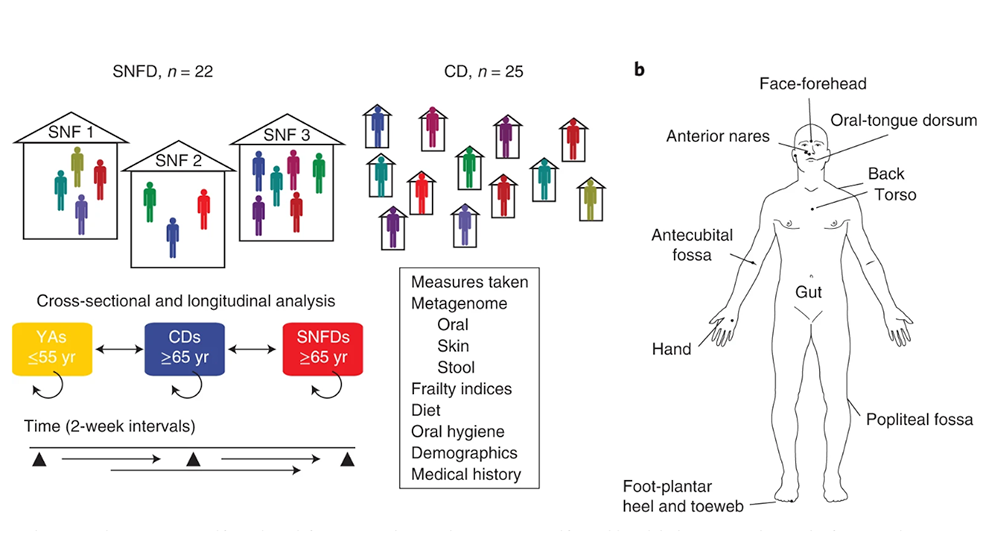
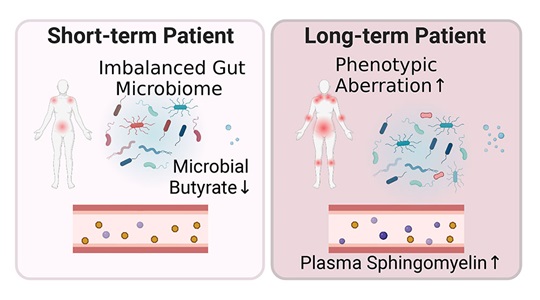
The Oh Lab
We seek to use diverse technologies like genomics, synthetic biology, and genome engineering to target and manipulate the microbiota for therapeutic purposes.
Principal Investigator
Location
Connect
Topics
Our research
The long-term goal of our research program is to create new microbiome-based therapeutics for skin disease, with implications for a wide range of inflammatory diseases. Our lab is dually computational and experimental - we develop advanced algorithms and analyses using shotgun metagenomic sequencing data for the purpose of reconstructing the structure and dynamics of microbial communities, and we complement our genomic predictions with in vitro and in vivo experiments to investigate gene function, visualize microbes, or test mechanism of action. Much of our work is focused on the human skin microbiome, and we also study the gut and airway microbiota to better understand systemic and local interactions of the microbiome with the immune system.
Our lab’s basic framework is to use our computational approaches to create highly informative maps of the fundamental characteristics of healthy vs. diseased microbiomes, to use cultivation, CRISPR, and experimental models to test our genomics-driven hypotheses and to define a phenotype for individual microbes, then finally, to identify and create strategies to engineer the microbiome for therapeutic purposes.
Our current major disease areas of interest include: aging, skin disease, infectious disease, skin cancer, immunotherapy response and immunotherapy-associated side effects, chronic fatigue syndrome and other pro-inflammatory diseases.
Some of our research projects that span these disease areas include:
Refining the Microbial Blueprint
- Algorithms for improving reference-based and de novo metagenomic characterizations
- Experimental technologies (single cell, in situ sequencing, high throughput phenotyping) for reconstructing microbiota and host-microbiome interactions
The Diseased and Healthy Microbiome
- The genetic architecture and strain diversity of skin commensal Staphylococcus epidermidis and its role in skin health and infectious disease
- Defining the host-microbiome immune interactome – large-scale reconstruction of the complex network linking microbial products to aberrant immune activation
- Local and systemic interactions of the gut and skin microbiome in skin cancer progression and immunotherapy response
Engineering the Microbiome: Therapeutics
- Engineering Staphylococcus epidermidis as a drug delivery chassis
- Defining the rules for engineering the skin microbiome
- Modeling engraftment of foreign microbiota and immune interactions in mice
In these projects, we use diverse technologies such as metagenomics, comparative genomics, single cell, culturomics, imaging, metabolomics, CRISPR, synthetic biology, metabolomics, and high-throughput screening.