
Albert Cheng, center, with Nathan Jillette and Menghan Du.
Transgenes are genes transferred from one organism to another, and they are important for many research areas. For example, it’s possible to introduce human versions of genes into mouse cells or even live mice to study their effect on the cells’ or animals’ phenotypes, or traits.
The primary challenge involved with transgenes is the actual delivery of them into the cells. To engineer cells, the most common method is to assemble the transgene and associated sequences on a bacterial plasmid, then use viral vectors to move the plasmids into the recipient cells. Recently, engineering a gene within the cells using What is CRISPR?CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats.CRISPR/Cas techniques is also possible. Neither technique is highly efficient, however, making it necessary to select the cells that actually receive a functional version of the desirable gene and discard the rest.
To determine which cells to keep, researchers use what are known as selectable markers. These are genes added to the plasmid that provide an easily identifiable trait, such as resistance to an antibiotic or fluorescence. Selectable markers work well for research involving single transgene, but adding more than one transgene to cells can be challenging to track. There are a limited number of fluorescence proteins, and subjecting cells to multiple antibiotic selections after each plasmid is delivered can harm them. To address the issue, JAX Assistant Professor Albert Cheng, Ph.D., led a team to develop split selectable markers, a method that can indicate the successful integration of multiple transgenes.
In “Split Selectable Markers,” a paper published in Nature Communications, Cheng et al present how split selectable markers are made and can be used. Simply put, the split markers involve dividing the marker genes at specific sites and fusing them to protein splicing elements. The divided fragments are then added to separate plasmids, each of which also includes a transgene. When both plasmids are successfully delivered into a cell, a full-length marker protein is produced via protein trans-splicing. Cells that only receive one plasmid will not express the full marker protein and can therefore be selected out via antibiotic treatment or identified via lack of fluorescence. Cells receiving both plasmids could be selected at very high percentages, generally more than 90% and up to 100%, for subsequent transgenic research.
The system is not limited to simple two-plasmid applications, and the researchers also present a protocol for selecting up to six transgenes using a single split selectable marker. In addition, split selectable markers can be used to facilitate genome engineering using the CRISPR/Cas system when it is used to “knock-in” (add) a genetic sequence at a specific location in the genome. Current precision editing and knock-in methods are inefficient, underscoring the need for effective marker and cell selection systems. Using split markers in concert with CRISPR/Cas editing again yielded very high percentages of cells with the desired genotypes following selection and growth.
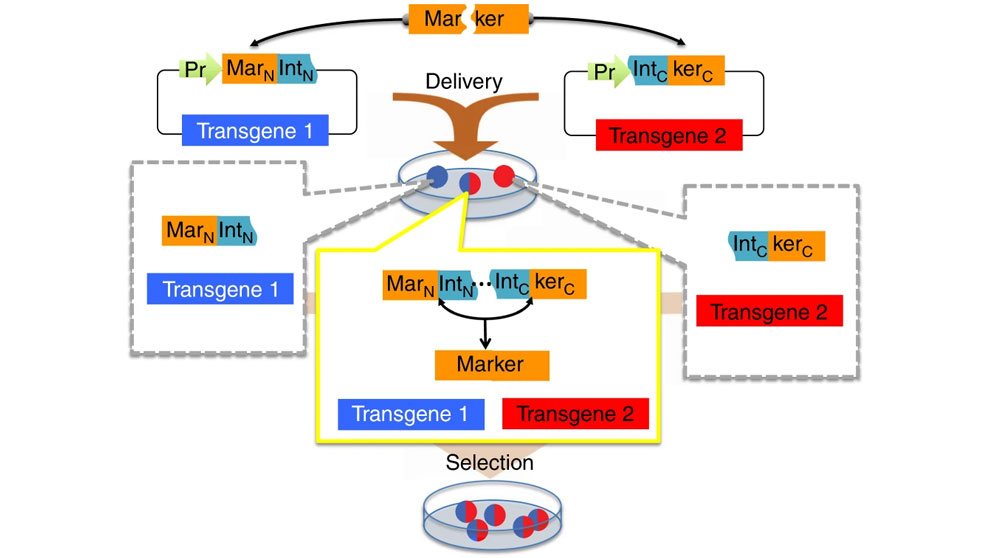 Split selectable marker for co-selection of two separate transgenic vectors. The coding sequence of a selectable marker is split into an N-terminal fragment (MarN) and a C-terminal fragment (kerC) and separately cloned upstream of an N-terminal fragment of a split intein (IntN) and downstream of a C-terminal fragment of the split intein (IntC), respectively, on two different vectors each carrying a different transgene. These vectors are delivered to cells yielding sub-populations of cells containing either one, or both of the vectors. Only cells with both vectors expressing the two intein-split selectable marker fragments (“markertrons”) undergo protein trans-splicing to reconstitute a full-length selectable marker, allowing specific selection and enrichment of the double transgenic cells.
Split selectable marker for co-selection of two separate transgenic vectors. The coding sequence of a selectable marker is split into an N-terminal fragment (MarN) and a C-terminal fragment (kerC) and separately cloned upstream of an N-terminal fragment of a split intein (IntN) and downstream of a C-terminal fragment of the split intein (IntC), respectively, on two different vectors each carrying a different transgene. These vectors are delivered to cells yielding sub-populations of cells containing either one, or both of the vectors. Only cells with both vectors expressing the two intein-split selectable marker fragments (“markertrons”) undergo protein trans-splicing to reconstitute a full-length selectable marker, allowing specific selection and enrichment of the double transgenic cells.