Natural genetic variation
I.A. QTL analysis of maximum life span in wild-derived inbred lines of mice
In our search to discover genes that regulate maximum life spans, we studied two 4-strain cross populations derived from a total of 8 mouse strains. Each 4-strain cross included one wild-derived strain and 3 diverse inbred strains. We found alleles at 2 loci (5–10 cM chromosome regions identified by genetic markers) that extend maximum life spans (the longest lived 16–40%, giving maximum power (Klebonov & Harrison, 2002) by at least 10–15% (Klebanov et al., 2001).
In the first cross, the longest-lived carriers of the D8Mit171 marker allele from the MOLD/Rk strain, Mus musculus molossinus, outlived the longest-lived noncarriers by more than 4 months. We identified this gene as Leg1 (longevity extending gene 1). In the second cross, the longest-lived carriers of the D10Mit267 allele from the CAST/Ei strain, Mus musculus castaneus, also outlived the longest-lived noncarriers by more than 4 months. We identified this gene as Leg2 (longevity extending gene 2). In both crosses, the effects were significant, and each effect required only one copy of the allele (indicating that the allele is either dominant or additive). Importantly, the single allele that extended life span was from the wild-derived strain in each of the crosses.
Because these life span increases require a delay in the aging of all essential biological systems, it is likely that these "longevity" alleles retard basic aging mechanisms in multiple biological systems simultaneously. These results led us in 2 directions: 1) We are conducting fine mapping and candidate gene analysis of Leg1 (Leg1a) and Leg2 (Leg2a). This process includes construction of congenic mice and studies on the effects of the genes in the context of various F1 hybrids. 2) Because the alleles that affect the increased life span came from the 2 wild-derived strains, we are conducting an extensive search for additional alleles using 4, new, wild-derived strains.
I.B. Biomarker analysis of aging in a wild-derived stock (Pohnpei mice)
One of our most exciting programs involves Pohnpei (Pohn) mice, a stock of mice (Mus musculus) that was trapped on the Pacific island of Pohnpei. Pohnpei has few murine predators, and evolutionary theory predicts that the expected reduction in ecological mortality will promote the evolution of retarded aging. In collaboration with Steven Austad and Richard Miller, we imported one female and 3 male Pohn mice into The Jackson Laboratory and bred 5 "families."
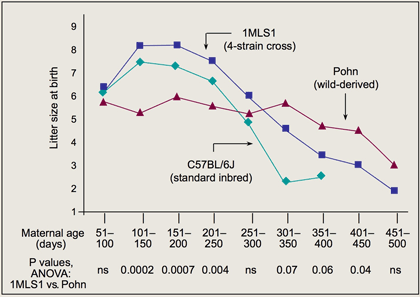
Although one family showed increased life span, in general, total life span is comparable to long-lived inbred strains. In all 5 families, however, female reproductive life span is greatly increased in comparison to laboratory (C57BL/6J [B6]) and 4-strain cross mice (Figure 1). This phenotype is associated with increased ovarian follicle numbers (Flurkey et al., 2007
).
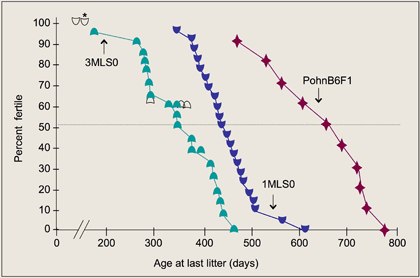
Furthermore, F1 hybrids between Pohn and B6 reproduce 5–6 months longer than do 4-strain cross populations (Figure 2) or other long-lived F1 hybrid mice (Flurkey et al., 2007).
The Pohn mice at The Jackson Laboratory are now fully inbred.
I.C. The Diversity Outcross (DO) Mice
In the search for genes that regulate aging and healthspan, our basic research strategy is to identify the genes that mediate the effects of experimental treatments on aging-related phenotypes and life span. By identifying the allelic distribution — comparing expression in short-lived vs. long-lived animals — we can thus identify specific alleles that delay aging and extend healthspan.
Recently, we started using a powerful new mouse resource — the Diversity Outcross (DO) stock — in our search for aging genes. The DO stock, designed by Dr. Gary Churchill at The Jackson Laboratory, is a novel heterogeneous stock of mice derived from 8 diverse parental strains. Five strains are classical inbred strains: 129S1/SvImJ, A/J, C57BL/6J, NOD/LtJ, NZO/HlLtJ. Three strains are wild-derived inbred strains from different Mus musculus subspecies: CAST/EiJ (M. m. castaneus), PWK/PhJ (M. m. musculus), and WSB/EiJ (M. m. domesticus). The 3 wild-derived strains contribute 75% of the genetic diversity of the DO stock, making it at least 4 times more genetically diverse than would be possible using only the standard classical inbred strains (Roberts et al., 2007).
The DO stock has 2 advantages especially relevant to the search for genes that regulate aging and aging-related phenotypes: First, because the DO is an advanced intercross, it has a high density of recombinations. This makes the DO more than 10 fold more precise than a standard F2 or N2 cross in candidate gene identification, allowing us to combine both traditional and fine mapping into one step, greatly accelerating gene discovery. Figure 3 shows the low levels of linkage disequilibrium in DO mice, which enable high-resolution mapping within just 10 generations of crossing. Second, because of the extreme genetic diversity of the DO stock, if we discover allelic variation that influences life span or other aging-related phenotypes, it is likely that such variation affects aging itself. In contrast, with a less diverse stock, such variation could simply have affected the risk of disease in just one or a few parental strains.
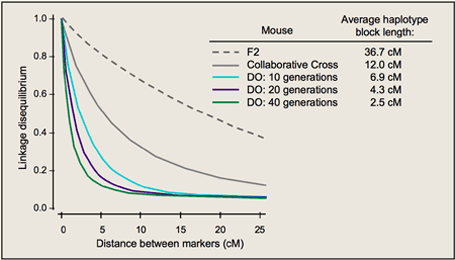
Simulation details: Outbreeding is by the current DO breeding scheme. Crossovers are calculated based on the Haldane model of recombination. Estimates are based on 100 replicates for each condition. Linkage disequilibrium is measured using a multi-allelic generalization of R2 that allows 8 alleles and collapses to R2 for the biallelic F2 cross.
Another important advantage of the DO stock is that we study natural allelic variation and distribution to identify structural alterations of regulatory pathways and gene targets. With genetically-engineered allelic variation, it would not be surprising for beneficial effects to be accompanied by unpredictable, serious, adverse effects. With natural allelic variation, however, this would be much less surprising — any allele with serious adverse affects would have been unlikely to survive generation after generation. Thus, any intervention that might result from a study using DO mice could be expected to induce fewer adverse side effects, making it more promising for clinical application.
Our use of the DO as a model system is also very exciting from the perspective of gerontological research in general. The DO is part of a complementary resource system that includes hundreds of Collaborative Cross (CC) recombinant inbred strains (derived from the same 8 parental strains as the DO stock) and their F1 hybrids (Churchill et al., 2004; Chesler et al., 2008). Using the information generated from studies of DO mice, the CC strains will provide a powerful means to explore the complex genetic interrelationships that govern aging, which will extend our work with DO mice far beyond our current research.