We are characterizing cre strains useful for craniofacial research as part of a related resource, the JAX Cre Repository. a centralized, comprehensive set of well-characterized Cre Driver lines and related information resources.
Strains listed below have undergone in-depth characterization in our group, following the Cre Repository staining protocols. Strains are listed alphabetically by promoter.
You can also visit creportal to search for Cre-expressing strains by promoter or site of activity, view detailed characterization of Cre activity including images, and get links to strains available in repositories.
Stock number(links to data sheet) | Strain name | Promoter (species) | Expected site of expression | Expression data thumbnail |
|---|---|---|---|---|
Krt17, keratin 17 (mouse) | Cre recombinase expression is driven by the endogenous keratin 17 (Krt17) promoter/enhancer elements |  | ||
Osr2, odd-skipped related 2 (Drosophila), mouse, laboratory | Cre recombinase activity is reported in developing palate and urogenital tract |  | ||
Trp63, transformation related protein 63 (mouse) | Cre recombinase expression is driven by Trp63promoter/enhancer elements | 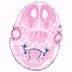 | ||
Prrx1, paired related homeobox 1 (rat) | Cre recombinase activity is expected in early limb bud mesenchyme and in a subset of craniofacial mesenchyme, along with limited female germline expression | 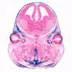 | ||
Tgfb3, transforming growth factor, beta 3 (mouse) | Cre recombinase activity is expected in the heart, pharyngeal arches, otic vesicle, mid brain, limb buds, midline palatal epithelium, and whisker follicles during embryo and fetus development |  | ||
Wnt1, wingless-related MMTV integration site 1 (mouse) | Cre recombinase activity is expected in embryonic neural tube, midbrain, caudal diencephalon, the mid-hindbrain junction, dorsal spinal cord, and neural crest cells |  | ||