An algorithm for optimally combining data from multiple structural variation detection methods
An algorithm for optimally combining data from multiple structural variation detection methods
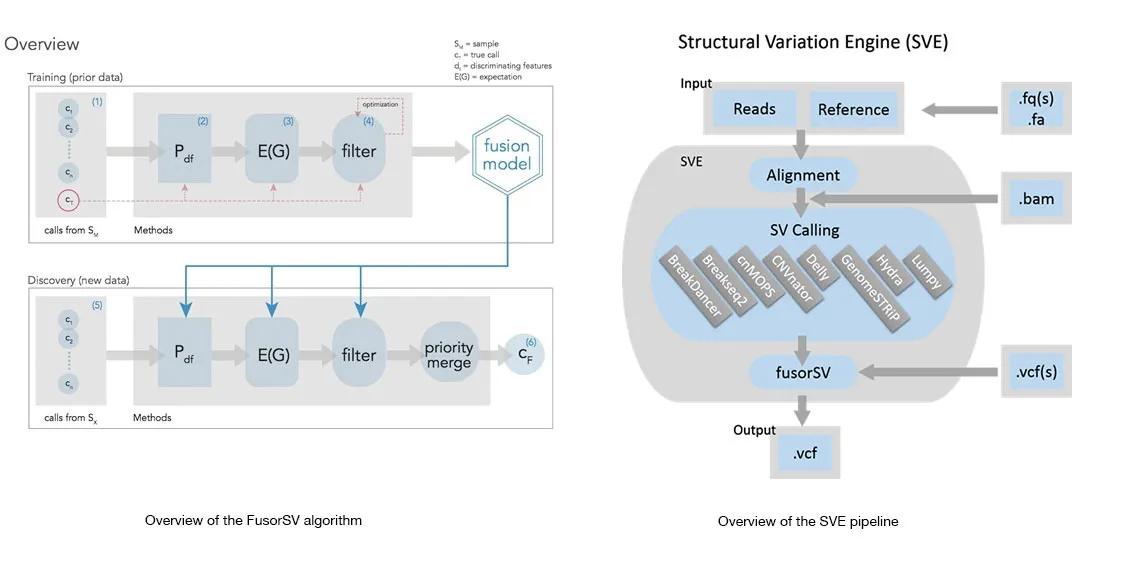

Comprehensive and accurate Structural Variation (SV) discovery from next generation sequencing data remains a major challenge. Popular approaches to overcome performance limitations of existing SV-calling algorithms are to use multiple complementary algorithms to determine the SV loci and then merge them under a heuristic manner. However, such approaches do not take into account the strengths and weaknesses of individual algorithms and hence either under or over merge the variant loci resulting in missing and/or false SV calls. Here, we present FusorSV, an open source tool that uses a data mining approach to assess performance and merge callsets from an ensemble of SV-calling algorithms. We also developed a FusorSV fusion model that was built on an ensemble of eight SV-calling algorithms and the analysis of 27 deep-coverage (50X) human genomes in the 1000 Genomes Project (1000GP). The model can be used for analysis of any newly sequenced sample and can be updated for other ensembles of SV-calling algorithms or datasets. Our model identified additional 843 (610 deletions, 202 duplications and 31 inversions) novel SV calls that were not reported by the 1000GP for the 27 samples. Experimental validation of a subset of these novel SV calls yielded a validation rate of 86.7%. For an easy-to-use SV detection pipeline, we built Structural Variation Engine (SVE) consisting of eight state-of-the-art SV-calling algorithms and FusorSV that is capable of performing gold standard SV analysis for whole genome sequencing projects that use Illumina paired-end read data.
Download and install from Github
Researchers at The Jackson Laboratory conduct pivotal study into aging and lifespan to uncover new details about how diets might make people live longer — but also their negative side effects.

Leading with compassion, Elizabeth Charnysh represents a team of genetic counselors established seven years ago as part of a comprehensive strategy to study the impact of genomic medicine research on human health.
View more
Genetic testing is a powerful tool in the fight against cancer, playing a crucial role in both prevention and personalized treatment. But what exactly are the different types of genetic tests, and how do they help?

Students arrive in Bar Harbor and Farmington as the SSP celebrates its 100th anniversary.