Overview: There may be several protocols listed on a strain datasheet because they are specific for different alleles. These alleles may be for a double or triple mutant strain, or they may be for mutations that are characteristic of the genetic background of the mouse strain.
JAX Example: Let's take a look at the 5xTG-AD Alzheimer's strain datasheet (Stock #006554, 34860-JAX) as an example. We can see that there are multiple genotyping protocols listed.
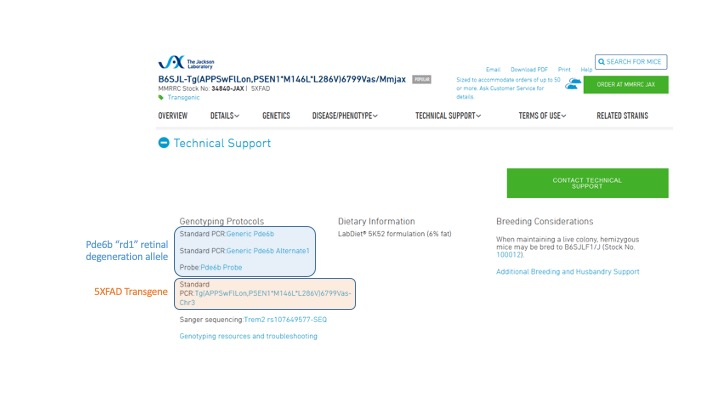
This strain is on a mixed genetic background of C57BL/6 and SJL. The SJL background expresses a retinal degeneration allele Pde6brd1 "rd1", which causes blindness when homozygous. Since the rd1 allele causes retinal degeneration when homozygous, it may be important to genotype. Several of the listed protocols are for the rd1 allele, and researchers can choose among these to genotype for the rd1 allele.
The standard PCR protocol for the 5xTg transgene is the only one that can genotype the 5xFAD allele. Since this is the allele that determines whether the mice will get Alzheimer's disease, you should use this protocol when breeding this strain.
How to Choose: Some strains have multiple alleles that you need to genotype, potentially including unwanted alleles from the genetic background of the strain. To choose which genotyping protocols(s) you need, start by determining which alleles you need to genotype, and select a protocol for each allele.
Overview: Sometimes there will be multiple protocols listed for the same allele, but there will be different protocols for different types of PCR. Some of the techniques that we use at JAX are Standard PCR, Melt Curve Analysis, QPCR, Probe/Endpoint, and sequencing. For an introduction to the different types of PCR, please review the articleHold the Agarose.
JAX Example: For this Green Fluorescent Protein (GFP) reporter strain there are three protocols listed – one is a standard PCR, one is a QPCR, and one is a probe-based assay. All three protocols are a generic GFP protocol that would work for any GFP-expressing strain.

The standard PCR and probe assays are not capable of differentiating hemizygous from homozygous animals. So, if you are breeding using a hemizygous x homozygous, or hemizygous x hemizygous breeding scheme, then multiple genotypes will be produced and you may need to use the QPCR to identify the genotype of the pups. In contrast, if you are breeding noncarrier x hemizygote you can use the standard PCR because homozygotes will not be produced, therefore you won't need to differentiate them from the hemizygotes using PCR.
How to Choose: There may be multiple protocols listed that are different types of PCR. Your choice of protocol in this scenario may depend on how you are breeding the strain and what type of PCR that you are capable of running in your facility. Start by determining what genotypes you need to differentiate, and choose a protocol that is capable of differentiating those genotypes.
Overview: Some strains may have multiple protocols listed for a single allele, and these protocols are different because of their specificity.
JAX Example: A good example of this is the Syn1-cre strain that has two genotyping protocols listed. There is a generic cre QPCR protocol that will work for virtually all strains that express cre. There is also a standard PCR protocol that is specific for this particular Syn1-cre. Your choice between these may depend on the breeding scheme that you are using. If you are breeding to homozygosity and need to differentiate hemizygous and homozygous animals, then you will need to use the generic Cre QPCR. If you intend to make a strain that expresses several cre alleles, or need to differentiate between two cre strains, then you should use the standard PCR protocol that is specific to this Syn1-cre.

Specificity information can be deduced from several locations on the protocol. First, the protocol name can be informative. Any protocol name that includes the term "generic" will work for multiple strains. Some examples that you will often encounter are "Generic GFP/YFP", "Generic Cre", and "Generic Neo". Another place to check for specificity is in the primer sequences. If the note on the primer indicates that it is a generic allele, then that likely means the protocol will work for multiple strains. Perhaps the most informative section to determine specificity is the "Other Strains" portion of the protocol, which lists all of the JAX strains that utilize this protocol. If many strains are listed, then this is a generic protocol.
How to Choose: When multiple protocols are listed for the same allele in a strain, it may be because some are specific and others are generic. Your choice of protocol may depend on what genotypes you need to differentiate, and whether you intend to breed the strain to any other strains.
Overview: There may be protocols listed for the same allele, and the same type of PCR – they only differ in the primer sequence. Both of these protocols will work, and are simply different versions that may anneal to different regions of the allele.
Example: Let's go back to the example from the 5xFAD strain (Stock #006554). You may have noticed that there were 2 standard PCR protocols listed for the Pde6brd1 "rd1" allele. If you open up those protocols, you'll see that the only difference between them is the primer sequence and the sizes of the resulting bands.

Multiple versions may be available because we will often test multiple primer sequences, or develop new ones as time goes by. If the primers work well, then we'll make the protocol publicly available so that researchers have multiple assays to work with.
How to Choose: There may be multiple protocols listed that appear to be very similar. You may wish to try the primers from each protocol to see which one works best with your particular PCR setup.
To choose which genotyping protocol(s) you need, you'll need to start by asking yourself these questions. There is no one-size-fits-all approach, so addressing these questions will help you determine what genotyping protocol(s) will suit your particular application.