Optogenetic mice have been revolutionizing neuroscience research since they burst onto the scene in the first decade of the 21st century. In combination with an ever-expanding collection of cre recombinase driver lines, neurobiologists are now able to selectively activate or inhibit specific neurons with light and to study how they modulate downstream neuron electrophysiology ex vivo or neurological behaviors in vivo. One shortcoming associated with existing optogenetic technology has been the difficulty in monitoring directly the electrophysiological outputs of the optogenetically-stimulated neurons. Although reporter strains that allow visual tracking of calcium concentrations or voltage changes are available, the former only serve as surrogates for neural activity, and the latter have not been spectrally distinct enough from the stimulatory optogenetic proteins to allow simultaneous optical stimulation and recording of the evoked electrical waveforms in the same neuron.
A new collection of optogenetic tools is changing all that. “Optopatch” and the related “Floxopatch” mice are now available from the JAX® Mice Repository. These novel strains allow both optogenetic stimulation and electrophysiological output monitoring in the same neurons. With these mice neuroscientists now can optically manipulate and monitor neural activity ex vivo and in vivo with greater precision.
The “Optopatch” construct: The key to simultaneous optogenetic stimulation and recording
“Optopatch” and “Optopatch2” (aka “Floxopatch”) mice are the brainchild of Dr. Adam E. Cohen and colleagues at Harvard University and the Howard Hughes Medical Institute. The key to these strains is the Optopatch construct. As illustrated in Figure 1, it contains both a near-infrared, Archaerhodopsin-derived voltage indicator, QuasAr2, and a blue-shifted channelrhodopsin, CheRiff. The two optogenetic proteins are expressed as a single polypeptide which self-cleaves (due to a P2A self-cleaving ribosome skip sequence) to release the separated proteins.
The spectral properties of the two optogenetic proteins are sufficiently distinct to allow activation of Optopatch-expressing neurons with blue light via CheRiff and simultaneous recording of the evoked action potentials via the near-infrared-sensitive QuasAr2 protein.
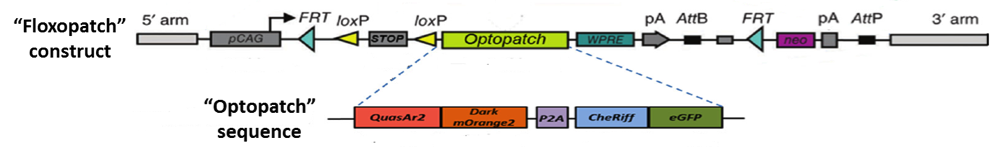
Figure 1: The “Optopatch” and “Floxopatch” constructs. The blue-shifted channelrhodopsin protein CheRiff is expressed as a fusion protein with an enhanced green fluorescent protein (eGFP) moiety that permits identifying cells that express the construct. Likewise, the QuasAr2 voltage sensor is expressed as a fusion protein with an mOrange2 fluorescent molecule. The latter, however, carries a mutation that renders it non-fluorescent. The “dark” mOrange2 protein facilitates transport of the QuasAr2 protein to the plasma membrane.
In Floxopatch mice (B6N.Cg-Gt(ROSA)26Sortm1Acoh/J - 028678), the Optopatch construct has been placed downstream of a cre recombinase-sensitive lox-stop-lox (LSL) cassette. Breeding Floxopatch mice to mice that express cre recombinase in specific neuron subtypes results in Optopatch protein expression in those neurons.
In their paper describing Floxopatch mice, the Cohen group bred them to several cre driver lines that expressed the recombinase in different neuron populations. Cre-dependent Optopatch protein expression did not affect the average body weights of the mice, nor did it affect the electrophysiological properties of dissociated target neurons in culture. Moreover, the action potentials recorded by fluorescence changes via the voltage-sensitive QuasAr2 protein mirrored those recorded by direct patch-clamp electrophysiological recording. In fast-spiking neurons, however, the amplitudes recorded via fluorescence tended to be lower than those recorded by patch clamp, indicating that improvements in the speed with which fluorescence-mediated reporters and detection systems operate will be necessary to accurately represent the electrical waveforms generated in the very fastest-firing neurons.
Dr. Cohen’s team also demonstrated that Floxopatch mice allow light-stimulated activation and fluorescence recording from the same neurons in cre-postive, LSL-Floxopatch (cre+, Floxopatch+) neurons in acute brain slices ex vivo and in peripheral neurons in vivo. In brain slices, the Cohen team observed that cre+, Floxopatch+ neurons could be stimulated repeatedly and their action potentials recorded with minimal change in amplitude or pattern over long intervals (at least 20 minutes). Stimulating and recording from the same neuron after such intervals is not as easy to do with conventional patch clamping techniques. The Optopatch measurements also revealed neurons in specific subpopulations with different firing patterns. These data suggest the neurons within a subpopulation may be divided into more refined categories based on their outputs. The Cohen team also optically stimulated and recorded from multiple neurons in intact jugular/petrosal/nodose ganglia (located in the neck) from Nav1.8-cre (Scn10a-cre) +, Floxopatch+ mice. The mice expressed cre/Floxopatch in primary nociceptive neurons of the trigeminal ganglia and dorsal root ganglia, and Optopatch stimulation and recording again revealed neurons with different firing patterns (Figure 2).
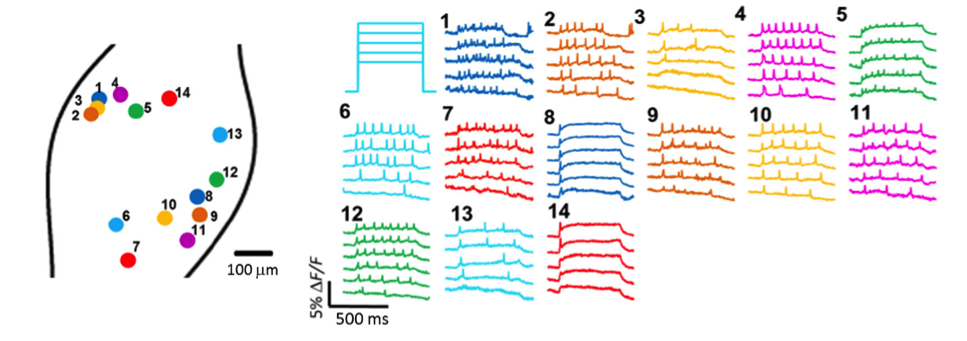
Figure 2: In vivo Optopatch imaging. Fluorescence recordings from 14 nodose ganglion neurons from Nav1.8-cre+, Floxopatch+ mice are illustrated. The neurons were stimulated using blue light with variable intensity ranging from 30-50 mW/cm2. Neurons with several different spiking patterns are shown.
JAX’s Optopatch/Floxopatch Collection
So far, three different Optopatch/Floxopatch strains have been deposited in the JAX® Mice Repository:
028678 B6N.Cg-Gt(ROSA)26Sortm1Acoh/J (Floxopatch)
029677 B6N.Cg-Igs7tm2(tetO-Optopatch2)Acoh/J (Ai141)
029679 B6N.Cg-Igs7tm2(tetO-Optopatc3,CAG-tTA)Acoh/J (Ai155)
The latter two strains combine both the cre- and tetracycline-inducible systems. Ai141 mice must be bred to both a cre-expressing and a tetracycline transactivator (tTA/rtTA)-expressing strain to express the Optopatch proteins. When Ai155 is bred to a cre-expressing mouse, the tet-OFF tTA protein is constitutively expressed and drives expression of the Optopatch proteins. (Doxycyline/tetracycline administration to this cre-recombined mouse actually represses Optopatch expression.)
In the nearly two decades since the “Decade of the Brain”, the explosion in sophisticated genetic tools at neurobiologists’ disposal has been staggering. Optopatch and Floxopatch mice are the latest and among the most sophisticated “wrenches” in the mouse neurobiologist’s toolbox. With an ever expanding collection of cre recombinase-expressing driver lines, cre-dependent and independent fluorescent reporters, such as “Brainbow" mice, and light-inducible optogenetic strains, neurobiologists can now map and control neuronal circuits with unprecedented precision as a means to better understand their characteristics and their influence on behavior and neurological disease.