Cre-lox myths busted
In a recently published paper (Heffner, et al. 2013) researchers at The Jackson Laboratory have found that many strains expressing Cre have phenotypes that were not obvious when these strains were created. Some of these phenotypes, including unexpected expression patterns, may seriously complicate the interpretation of your experiments.
Here we dispel four common Cre-lox myths:
Myth #1. Cre strains have excision activity only in the intended cell type or tissue.
You expect that a strain where Cre is driven by a brain-specific promoter will express Cre only in the brain. Think again! The majority of the strains that we have characterized show unexpected Cre activity in off-target tissues. An example of this is strainB6.129S2-Emx1 /J (005628), which expresses Cre in the kidney and thymus, in addition to the brain.
Myth #2. All Cre strains have predictable deletion efficiencies.
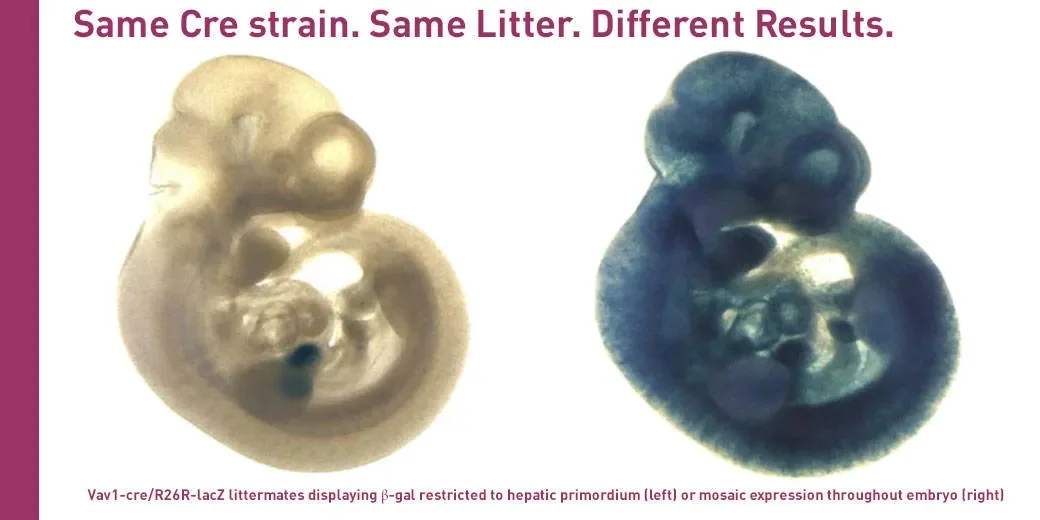
Not all of them do. In several Cre-expressing strains, for example B6.Cg-Tg(Vav1-cre)A2Kio/J (008610) and B6.Cg-Tg(Fabp4-cre)1Rev/J (005069), Cre expression may be quite variable even among litter-mates as shown in the picture above. Watch out for strains that show mosaic patterns of Cre expression, too. If you are working with such strains you may find quite a bit of variability, and it may be harder to reproduce your results. Often you may need to analyze the phenotypes of more mice before you can reach meaningful conclusions.
Myth #3. Recombination efficiency of Cre is the same whether the male or the female parent supplies the transgene.
We have found that for most models that we have characterized using aCre-reporter strain, the pattern of Cre expression is independent of which parent passes on the Cre transgene. With certain strains, however, Cre recombinase activity is more robust in offspring that inherit the Cre from their mother compared to those that inherit Cre from their father. Examples of this phenomenon are seen in strains carrying the EIIa-Cre transgene (stains003314 and 003724).
Myth #4. Strains only expressing the Cre transgene have no adverse or observable phenotypes.
It has been well documented that Cre expression in mouse models can have toxic side effects such as infertility and decreased viability. This is especially true for certain strains when they are homozygous for the transgene. High levels of Cre activity may result in recombination between cryptic loxP sites in the genome, resulting in deletions or translocations that may result in reduced viability. Also, for Cre strains created by microinjection, the transgene’s insertion into the genome may disrupt the function of endogenous genes that are critical for the viability and fertility of the animals. Examples of Cre transgenic strains that are not viable as homozygotes include B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J (004682) and B6.Cg-Tg(CD2-cre)4Kio/J (008520).
Have I burst your bubble? You may be asking yourself what to do next. Before you go on to generate your next tissue- or developmental stage-specific conditional knock-out, make sure that you are aware of the pattern of Cre expression of the strain you will be using. TheJAX Cre resource has characterized the expression patterns of over 50 Cre-expressing strains, with more data to come. Be an informed consumer and check it out! If Cre expression data for a particular strain is not publicly available, we encourage you to generate it in your lab.
A recent article discusses The Jackson Laboratory’s (JAX’s) high-throughput Cre expression characterization pipeline and JAX resources that can facilitate research using Cre-lox technology. In the long run, this information will enable you to interpret your results wisely and to better understand the function of your candidate gene in health and disease.
