Genes essential to life are enriched for human disease genes, large-scale mouse phenotyping study shows
Article | September 14, 2016
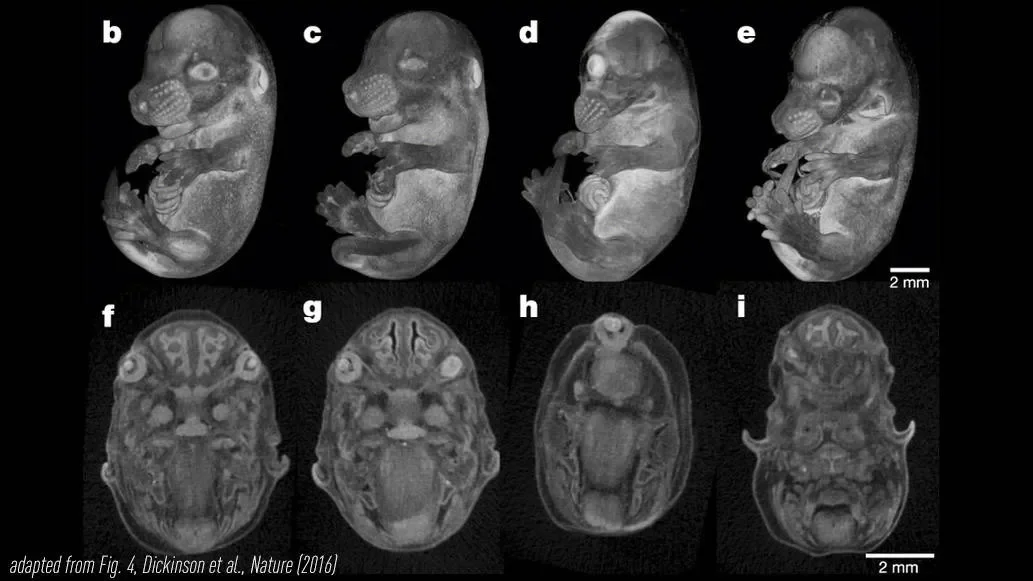
JAX is a member of the International Mouse Phenotyping Consortium, which is generating and phenotyping (assessing the morphological and physiological characteristics of) knockout mutations for all of the protein-coding genes in the mouse genome. The Consortium aims to discover new functions for the roughly 20,000 genes shared with humans, and makes all of the mouse strains available to provide a platform for dissecting the mechanisms of human disease.
This work is an enormous boon of information for researchers.
The Nature study, published Sept. 14, reports the results of the first 1,751 genes characterized by the IMPC, including the finding that nearly one third are essential for life. This includes 410 lines that are fully lethal (in other words, a mouse embryo missing any one of these genes fails to survive), and an additional 198 for which fewer than half of the expected number of embryos survive.
Using a new, standardized phenotyping pipeline and mouse strains on a single C57BL/6N genetic background, the researchers established both the time of embryo death and the nature of the lethal phenotypes for these lines, discovering many novel phenotypes that shed light on the function of these genes.
Study co-author Mark Henkelman, Ph.D., director of the Mouse Imaging Centre of the University of Toronto-affiliated Hospital for Sick Children, comments, “What sets this study apart is the use of high-throughput 3D imaging with automated, computational analysis to identify novel phenotypes that would have easily been missed by gross inspection. The results are striking and are surely going to set a new standard for the field.”
The team also showed that identification of essential genes in the mouse provides a window on human disease, including the discovery of a number of novel cases where human disease genes overlap with essential genes. In addition, in collaboration with the ExAC Consortium, they showed that human orthologs of these essential genes are significantly depleted for loss-of-function mutations, and that these genes are thus strong candidates for undiagnosed human genetic conditions.
“The work of the consortium will contribute significantly to our understanding of the genetic bases for human diseases, including spina bifida and cardiovascular defects, amongst many others,” said co-first author Dr. Lydia Teboul, head of molecular and cellular biology at MRC Harwell Institute, in the UK.
Moreover, notes JAX Senior Research Scientist Steve Murray, Ph.D., corresponding author of the study, “when looking across the seven or eight embryos generated for each knockout, we found variations in phenotype at a surprising frequency. We expect diversity when we look across different genetic backgrounds, but this is the first large-scale documentation of pervasive variable expressivity in a defined genetic background.”
“This paper is really focused on defining the phenotypes associated with genes that are essential to embryonic and post-natal development,” says co-first author Mary Dickinson, Ph.D., professor and Kyle and Josephine Morrow Endowed Chair of Molecular Physiology and Biophysics at Baylor College of Medicine. “This group of genes is particularly exciting because many that are essential in mouse are also linked to diseases in humans, and the paper reports the efforts to build a catalog of these findings for the community, complete with 3D images of the defects that were observed.”
All data and images generated by the project are available to the research community, disseminated via an open-source web portal in real time without embargo. The mouse models generated are also available to other researchers who may be investigating particular pathways or disease phenotypes.
“This freely available and accessible dataset provides significant new gene-phenotype associations to enable scientists to prioritize gene candidates identified in their preclinical and discovery research,” says co-first author Dr. Ann Flenniken, manager of the Clinical Phenotyping Core at The Centre for Phenogenomics in Toronto.
Co-author Maja Bucan, Ph.D., professor of genetics at the Perelman School of Medicine at the University of Pennsylvania, says, “The sheer amount of new data reported in this paper is impressive. We compared the genes analyzed in this paper with a list of known human disease genes, which made it possible to identify for the first time the mutant phenotypes in the mouse for 52 human disease genes.” Xiao Ji, M.S., co-first author and a doctoral candidate in the Graduate Group in Genomics and Computational Biology at Perelman, adds, “The IMPC effort has provided an unparalleled model organism resource for functional genomics studies.”
“This paper is just the tip of the iceberg,” said Dickinson. “We want the scientific community to know even more about IMPC efforts and that they have access to the mice as well as the phenotype data. This work is an enormous boon of information for researchers.”
This work was supported by the National Institutes of Health’s National Human Genome Research Institute and the Office of the Director’s Common Fund (U42 OD011185, U54 HG006332, U54 HG006348-S1 and OD011174, HG006364-03S1 and U42 OD011175, U54 HG006370). Additional support was provided by The Wellcome Trust, Medical Research Council Strategic Award, Government of Canada through Genome Canada and Ontario Genomics (OGI-051), Wellcome Trust Strategic Award “Deciphering the Mechanisms of Developmental Disorders (DMDD)” (WT100160), National Centre for Scientific Research (CNRS), the French National Institute of Health and Medical Research (INSERM), the University of Strasbourg (UDS), the “Centre Européen de Recherche en Biologie et en Médecine”, the “Agence Nationale de la Recherche” under the frame programme “Investissements d’Avenir” labeled ANR-10-IDEX-0002-02, ANR-10-INBS- 07 PHENOMIN, The German Federal Ministry of Education and Research by Infrafrontier grant 01KX1012.
Dickinson et al.: High throughput discovery of novel developmental phenotypes. Nature, Sept. 14, 2016, http://www.nature.com/nature/journal/vaop/ncurrent/full/nature19356.html.


