Defining the cellular targets for Huntington’s disease pathogenesis
A new publication in Nature Medicine (Wang et al. 2014) reports on the expression of mutant huntingtin protein (mHTT) in specific regions of the brain and defines the cellular targets for optimal therapeutic intervention. Huntington’s disease is caused by expression of a mutant huntingtin protein containing an elongated stretch of polyglutamine residues. While the mutant protein is ubiquitously expressed, only certain cells in specific regions of the brain undergo neurodegeneration—specifically cortical and striatal neurons. The new findings indicate that therapies will need to target these cell types for optimal treatment.
Targeted reduction of mutant huntingtin protein expression
The research team at UCLA took advantage of the Cre-lox system to create mice with reduced expression of human mHTT in specific regions of the brain. This approach allowed them to define regions of the brain most influential in the pathogenesis of disease and thus the most important for therapeutic targeting. The key animal used in this approach was the BACHD [FVB/N-Tg(HTT*97Q)IXwy/J, 008197] strain. The bacterial artificial chromosome construct in this strain contains human mHTT with exon 1 flanked by loxP sites. Breeding to a Cre-expressing strain deletes exon 1, diminishing mutant protein expression. The authors used two different Cre-expressing strains in this study. The Emx1-cre transgenic strain utilizes a promoter to drive Cre expression primarily in cortical pyramidal neurons (CPN). They also used an Rgs9-cre transgenic strain to drive Cre expression primarily in striatal medium spiny neurons (MSN). Specific regional expression was validated by breeding these Cre lines, either singularly or together to create double transgenics, to two different reporter strains. The first was B6;129S4-Gt(ROSA)26Sortm1Sor/J (003309) which expresses lacZ in Cre expressing cells. The second strain was B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J (007903), a yellow fluorescent protein reporter. Following histological validation of brain region-specific expression, the authors bred the Cre transgenics to BACHD to create three cohorts of mice. Line BE (BACHD; Emx1-cre), line BR (BACHD; Rgs9-cre ) and BER (BACHD; Emx1-cre; Rgs9-cre).
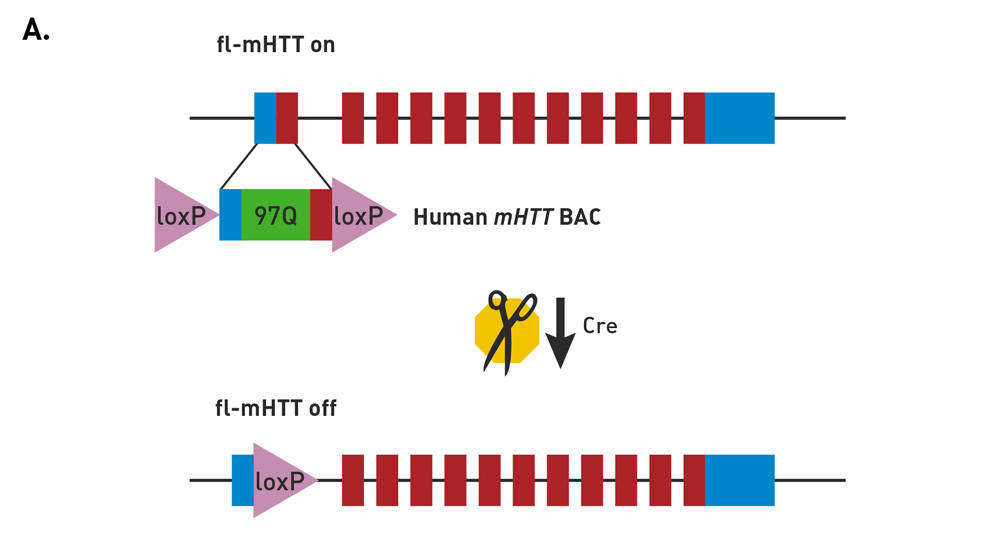
Molecular construct of the BACHD mice
FVB/N-Tg(HTT*97Q)IXwy/J mice (strain 008197, commonly called BACHD) express a neuropathogenic, full-length human mutant Huntingtin (fl-mhtt) gene modified to harbor a loxP-flanked human mutant htt exon 1 sequence (containing 97 mixed CAA-CAG repeats encoding a continuous polyglutamine (polyQ) stretch). Breeding to a Cre-expressing strain enables cell-specific reduction in mutant protein expression.
Defining the roles of cortical and striatal mutant protein expression
The authors quantified the regional reduction in mutant HTT protein expression by in situ hybridization and western blots. Compared to BACHD mice, BE mice showed 70% reduction in cortex and 40% reduction in striatum. BR mice showed a 44% reduction in striatum, and the BER mice showed 80% reduction in both cortex and striatum with no reduction in cerebellum. Motor performance was measured using the accelerating rotorod test and spontaneous locomotion by using an automated open-field test. BE mice showed greater improvement in both rotorod and open-field performance than BR mice, whereas the BER mice showed the greatest overall improved performance. Psychiatric-like behaviors for anxiety and depression were also examined using light-dark chambers and force-swim assays, respectively. Both BE and BER mice showed significant improvement while the BR remained essentially unchanged.
The authors also investigated the influence of region specific reduction of mHTT expression on corticostriatal synaptic communication, a known characteristic of HD. The BACHD mice had reduced expression of the presynaptic protein synaptophysin and the postsynaptic proteins actinin-2 and postsynaptic density-95—markers for synaptic neurons with roles implicated in receptor signaling dysfunction. Interestingly, their results suggested that reduced mHTT expression in the CPN (BE mice) not only led to improved pre and postsynaptic marker expression in the genetically targeted cells (cell autonomous correction), but also improved postsynaptic marker expression in the non-genetically targeted cells (non-cell-autonomous correction) of the MSN. In BER mice, all three markers were corrected and were not significantly different than wild-type mice.
Targeting both cortical and striatal neurons
Interestingly, the reduction of mHTT in the CPN (BE mice) significantly improved certain aspects of both motor function and psychiatric-like behavioral deficits (anxiety and depression). However, the BE mice still retained evidence of neurodegeneration as indicated by decreased forebrain weight, cortical volume loss and striatal volume loss. Targeting only the MSN (BR mice) demonstrated the least improvement in motor function, psychiatric-like behaviors, and neurodegeneration. Targeting both regions of the brain, cortical and striatal neurons, led to the greatest overall improvement in all of the measured phenotypes. Therefore, these studies provide valuable insight into the regions of the brain requiring reduction of mutant huntingtin protein and set the stage for developing the most appropriate targeted therapy.
Additional research resources available for Huntington’s disease research:
Explore JAX resources for neurobiology research.
Download the field guide to working with mouse models of Huntington’s disease.
Obtain mice from The Jackson Laboratory Cre Repository.
pharmacology services for neuromuscular and neurodegenerative disordersin vivo Take advantage of .