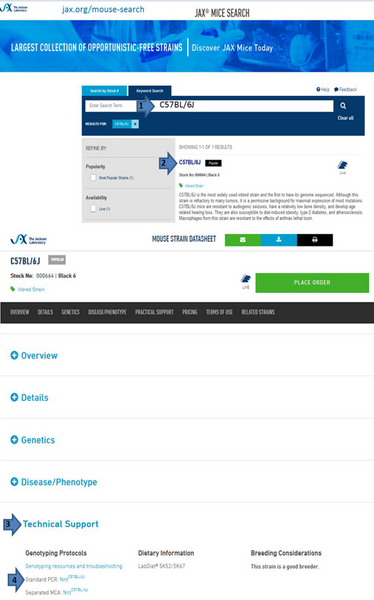
Access JAX® Mice genotyping protocols
There is an easy way to locate genotyping protocols for a specific strain on our website.
- Find the data sheet of a strain of interest by stock number (catalog number), by gene name or by strain name.
- Select the relevant strain from the list that shows up.
- Scroll down and click on “Technical Support”. A list of available genotyping protocols shows up.
- Select the relevant link to see the full protocol.