The JAX Clinical Knowledgebase (CKB) is a valuable resource that can be utilized to interpret somatic variants and identify the most relevant therapy via efficacy evidence annotations. Here is a list of the most common questions received from new and current users worldwide.
1. I already use a commercial vendor for my clinical reporting. What is the advantage to also using a knowledgebase such as CKB?
- The content used to generate a clinical report includes only a snapshot of the data collected in the context of the patient and their genomic profile. A knowledgebase—like CKB—can be used to access up-to-date data for confirmatory purposes and to provide a more holistic view of the data. For example, for a particular gene variant, it may be worthwhile to explore other variants in close proximity, providing a better understanding of how alterations within a specific domain or region affect the activity of the protein. In addition, there may be interest in identifying therapeutic efficacy evidence related to the gene variant that is not present in the clinical report.
2. Other than missense variants, what other data can I find using CKB?
- With CKB—including Core and Boost versions, you can find content specific to many other types of alterations, such as nonsense variants, frameshifts, in-frame deletions and/or insertions, duplications, splice variants, and fusions. CKB also includes content related to overexpression, decreased expression, and copy number variants (CNV), which are represented as either amplification, deletion, or loss.
Variants found in CKB align to the human reference genome GRCh38. Each variant within the database includes a table on the "Gene Variant Detail" page listing the reference transcript, the gDNA and cDNA coordinates, the corresponding protein change, the source database, and genome build.
3. What is the best web browser to use with CKB?
- We recommend using Firefox, but other browsers that work well include Chrome and Safari, but not Internet Explorer. The application also resizes for viewing capability on a mobile device, but we recommend using a computer for the best experience.
4. How can I recommend or request new data or a gene for CKB?
- Either email ckbsupport@jax.org when searching CKB, or if using either CORE or BOOST, click the green button (as shown in the figure below) in the top right-hand corner of various pages. This button reveals a drop-down box which includes the email link. This email address can be used to request new curation of genes, variants, and/or PubMed publications.

To request or recommend data while using CKB, click this green button to reveal the email link.
5. Is CKB data downloadable?
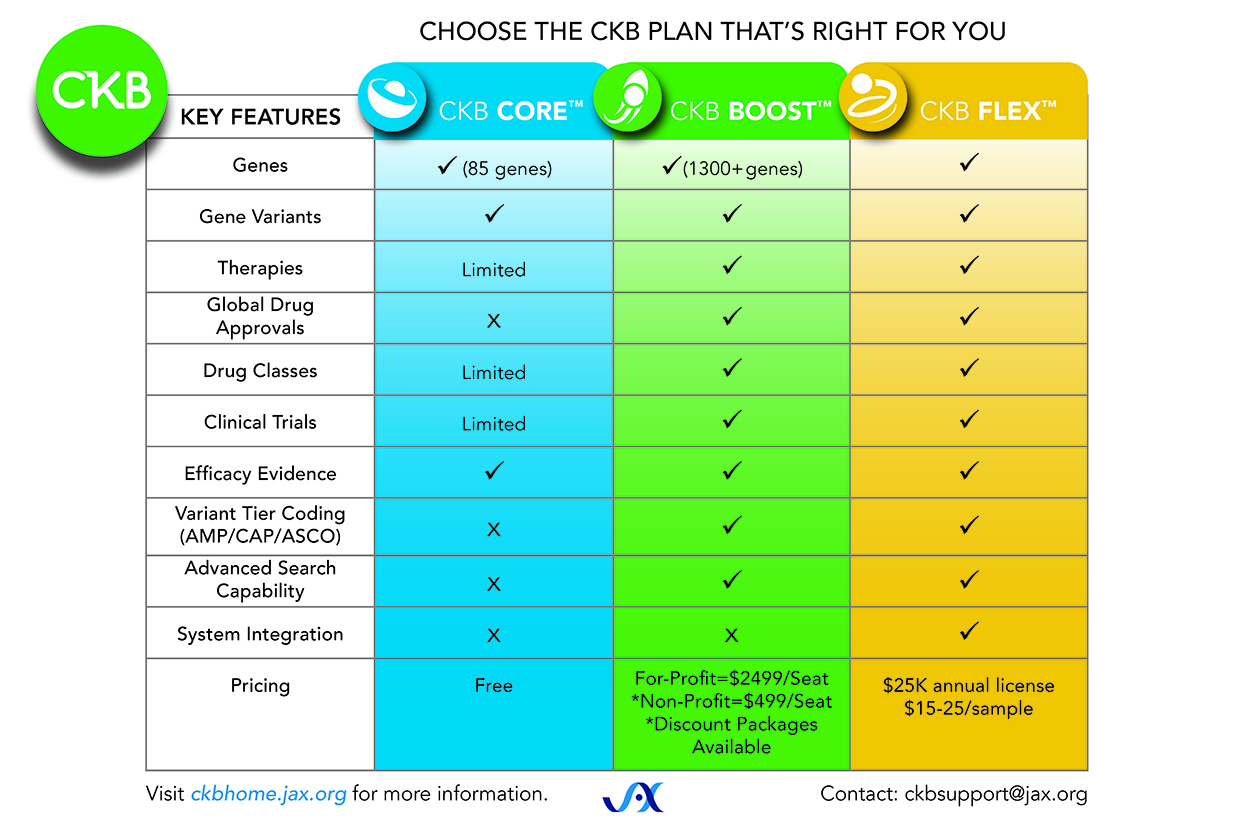
- Downloadable data from CKB is available using the FLEX version. CKB FLEX delivers CKB content as either a flat file or JSON, enabling CKB data integration. The product matrix below describes the differences and pricing between the three versions (CORE, BOOST, and FLEX).
6. Why are there some genes that include variants such as EGFR mut or BRAF act mut?
- CKB includes content that links variants to efficacy evidence and clinical trials. In some cases, there may be clinical trials or published studies in which the specific variants within a gene have not been identified or provided. For example, a study may note that patients within a cohort all harbor an EGFR mutation, but the specific mutations are not provided. Therefore, we added category variants, which allows us to accurately annotate the efficacy evidence. Below is a table listing the types of category variants that are included in CKB.
| Variant | Definition |
|---|---|
| act mut | Indicates that the variant results in a gain of protein function. This can be considered a category containing any variant that results in a gain of function. |
| fusion | Indicates a fusion of the gene, but the fusion partner is unspecified. This can be considered a category containing any fusion in which the specified gene is a partner. |
| inact mut | Indicates that the variant results in a loss of protein function. This can be considered a category containing any variant that results in a loss of function. |
| mutant | Indicates an unspecified mutation in the gene. This can be considered a category containing any missense, indel, nonsense, or frameshift variant in the gene. |
| rearrange | Indicates an unspecified rearrangement of the gene. This can be considered a category containing rearrangement of the specified gene. |
| exonX | Indicates a mutation in the specified exon. This can be considered a category containing any variant within the specified exon. |
| Xaa#(X) | Indicates that the variant results in the replacement of the amino acid at the specified position by another amino acid. The can be considered a category containing any variant resulting in an amino acid substitution at the specified position. An example would be ‘BRAF V600X’. |
| exon X del | Indicates a deletion in the specified exon of the gene. This can be considered a category containing any variant resulting from a deletion in the specified exon. An example would be ‘EGFR exon 19 del’. |
| exon X ins | Indicates an insertion in the specified exon of the gene. This can be considered a category containing any variant resulting from an insertion in the specified exon. An example would be ‘FLT3 exon 14 ins’. |
| del exonX | Indicates a deletion of the entirety of the specified exon. This can be considered a category containing any variant that results in the deletion of the specified exon, but not resulting in deletion of other exons. An example would be ‘MET del exon14’. |
7. What does it mean if the molecular profile for an efficacy evidence annotation has more than one variant?
- Molecular profiles can consist of one or more gene variants, and are linked to efficacy evidence annotations within CKB. Often, the efficacy of a drug is assessed in a cell line, patient tumor sample, or in an in vivo model that has more than one gene variant. Therefore, the molecular profile captures complex genomic signatures, providing a more accurate representation of the data. Complex molecular profiles may consist of any combination of the various types of gene alterations present in CKB.
8. Who does the curation for CKB?
- All curators of CKB are part of JAX and have either a Ph.D. or MD/Ph.D. They have extensive experience in assessing experimental data such as interpretation of graphs and figures and identifying whether appropriate controls were used. This scientific rigor ensures the integrity of the data within CKB. Furthermore, the complexity of cancer requires in-depth subject matter expertise in order to assimilate interpretations across heterogenous data sets and often discrepant data to draw quality conclusions.
9. What are the criteria used to assign a protein effect for a biologically characterized variant?
- The variant protein effect is based on the effect that the alteration has on the intrinsic activity of the protein, and is not centered on downstream pathway activation. Some variants that lead to a loss of intrinsic protein activity may result in activation of the pathway, but these variants would be assigned “loss of function” in CKB. For example, a common variant in KRAS, G12C, results in decreased KRAS GTPase activity leading to activation of downstream signaling. In CKB, we’ve assigned this the protein effect of “loss of function” due to the loss of the intrinsic activity of the protein, even though it results in pathway activation. The criteria used to assign protein effect can be found in the table below:
Categories of Functional Significance Description Gain of Function (Activating) Peer-reviewed published literature demonstrating functional evidence that the gene alteration results in increased intrinsic activity of the protein. Gain of Function-Predicted (Inferred Activating) The specific gene alteration has a location similar to other alterations in the same gene that have been functionally characterized as a gain of function within peer-reviewed published literature. Loss of Function (inactivating) Peer-reviewed published literature demonstrating functional evidence that the gene alteration results in decreased intrinsic activity of the protein. Loss of Function-Predicted (Inferred Inactivating) The specific gene alteration has a location similar to other alterations in the same gene that have been functionally characterized as a loss of function within peer-reviewed published literature. If all known functional domains or a key domain are lost (i.e. protein kinase domain), this interpretation is also appropriate. No Effect (Benign) Peer-reviewed published literature demonstrating functional evidence that the gene alteration results in activity similar to that of wild-type and does not demonstrate tumorigenic attributes. No Effect-Predicted Peer-reviewed published literature demonstrating modest functional evidence that the gene alteration results in activity similar to that of wild-type and does not demonstrate tumorigenic attributes. Unknown There is no peer-reviewed published literature demonstrating the gene alteration effects the intrinsic activity of the protein.
10. How are drug names decided?
- Drugs in CKB include those that are in either preclinical and clinical development or those approved for use in patients. The drug name is typically selected based on the name indicated in the reference used to describe the drug. Potential references include indexed publications, abstracts from professional meetings, or the NCI Drug Dictionary. Additional curation of a drug may include synonyms as well as trade name if approved. Drugs may also be assigned a drug class, which is dependent on the target of the drug.
If you have specific questions about using CKB or any of the available versions, please visit the JAX CKB homepage to learn more.
Recommended Resources:
CKB, powered by JAX, oncology spotlight: Revealing complex clinical genomic associations
Introducing: The Clinical Knowledgebase (CKB), Powered by The Jackson Laboratory