Light microscopy has been an immensely valuable tool in biology for hundreds of years. But as techniques and resolution improved, scientists ran into a problem based on the nature of light itself: its wave nature limits resolution to about 200 nanometers (nm), and anything smaller, such as sub-cellular structures and protein assemblies, could not be resolved. Electron microscopy far surpassed this resolution limit, but because of the conditions needed to perform it (sample fixation, which can introduce damage, vacuum conditions, etc.), many investigations stood to greatly benefit from higher resolution light microscopy.
Biophysicists have addressed the problem in ingenious ways, developing super-resolution fluorescence microscopy that moved light microscopy beyond the diffraction limit. Refined versions of this method use specialized fluorescent labels and dual-objective geometry to achieve many-fold improvements in resolution, in some cases approaching 10 nm in one focal plane. An important limitation remained however, in that samples could be a maximum of 700-1,000 nm thick. Mammalian cells are generally 5-10 microns thick, however (at least 5x thicker than allowed), and organelles that extended over multiple microns within the cell could not be imaged.
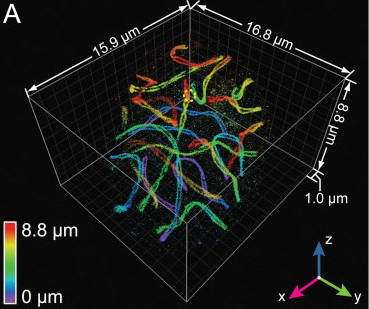
In a paper recently published in Cell, researchers led by Joerg Bewersdorf, Ph.D., of Yale University (formerly of JAX) and including JAX Senior Research Scientist Mary Ann Handel, Ph.D., developed and implemented a new microscopy method that allows resolution of three-dimensional structures to 10-20 nm throughout entire cells, in samples up to 10 microns thick. The method, called whole-cell 4Pi single-molecule switching nanoscopy (W-4PiSMSN), builds upon previous work by employing deformable mirrors that allow correction for aberrations introduced by sample depth. In addition, a new analysis algorithm provides automatic adaptation to changes in the shape and interference pattern of the point spread function, which is the response of the imaging system to the object being imaged.
In the paper, the authors demonstrate W-4PiSMSN capability on several difficult-to-visualize subjects, including networks of endoplasmic reticulum, cellular microtubules, mitochondrial networks, pore complexes on the nuclear envelope in the middle of cells, ciliary membranes and synaptonemal complexes in mouse spermatocytes during meiosis, which are protein complexes thought to mediate chromosome pairing and recombination during meiosis.
“We say that homologous chromosomes ‘find each other’ during recombination,” says Handel, who studies spermatogenesis and meiosis. “But the chromosomes are hugely complex structures containing immense amounts of DNA and protein, so having the right sequences bump into each other by chance would be pretty unlikely. Think about it — how do they do it? With microscopy at this resolution, we hope to visualize steps in the process, which would be amazing and help us understand a vitally important biological mechanism that is still unknown.”
The researchers also resolved T7 bacteriophages, showing their ~60-nm diameter capsid structure and pentagonal cross-section. They constructed 3-D movies of the structures as well, clearly showing such things as the complex topography of the nuclear envelope. In conclusion, the authors state that W-4PiSMSN “establishes 3-D biological imaging with molecular specificity and resolution in the 10-nm range as a general imaging technique.”
Huang F et al. 2016. Ultra High Resolution 3-D Imaging of Whole Cells. http://dx.doi.org/10.1016/j.cell.2016.06.016