The importance of understanding substrains in the genomic age
JAX Notes | September 30, 2003The importance of mouse models in biomedical research is well known and appreciated. The mouse is widely used in all types of research, including basic genetics, physiology, drug discovery and other aspects of modeling human disease. Genetically defined inbred mice are universally accepted as the principal model for analyzing and understanding inherited human disorders1-4.
Scientists use inbred strains because they are well characterized, and many commonly used substrains have been inbred for more than 200 generations. Except for gender differences, inbred strains can be regarded as genetically and phenotypically uniform, giving researchers the freedom to make comparisons and draw conclusions from a variety of experiments conducted around the world. In the current age of genomics, with the ability to genetically manipulate the mouse genome in an ever-increasing variety of ways, it is more important than ever to understand the differences among inbred strains and substrains of mice.
Most commonly used inbred strains, defined by 20 or more generations of consecutive sibling matings, were generated during the early 20th century. Early mouse geneticists readily distributed these inbred parent strains to colleagues around the world, who subsequently bred and distributed them to their colleagues. The result was the creation of multiple substrains within an inbred strain that became increasingly more genetically divergent through generations of independent inbreeding5,6.
As demand for commonly used inbred strains increased, mouse vendors established colonies of specific substrains and implemented various genetic quality-control measures to ensure the genetic integrity of their inbred strains. To the uninitiated user of the inbred laboratory mouse, the differences between individual substrains and, thus, differences between mice from different sources may seem unimportant. Upon closer scrutiny, substrain differences, primarily genetic in nature, could lead to inconsistent data, misinterpretation of results or even complete failure of an experiment.
Genetic differences among substrains can occur by several mechanisms:
- Residual heterozygosity at the time of separation.
- Undetected spontaneous mutations that become fixed in the colony. For example, a deletion of the alpha-synuclein locus has been observed in a substrain of C57BL called C57BL/6JOlaHsd7. In another case, two substrains of C57BL, C57BL/6JNmg and C57BL/6JKun, have shown phenotypic differences, demonstrating drift from each other, as well as from the C57BL/6J founder line8,9. While some investigators may think that genetic drift will not have a significant impact on their research, these examples show there may be severe consequences.
- Undetected genetic contamination or deliberate outcrossing of strains for specific experimental purposes10,11.
Substrains occur when colonies are separated by more than 20 generations from the progenitor strain, and are considered to be genetically distinct (refer to nomenclature guidelines at:www.informatics.jax.org/nomen).
Although it is often difficult to decipher the exact genetic causes, substrains from different sources can lead to functional differences among mice. For example:
- Behavioral differences among substrains12-14.
- Tissue rejection among related substrains (e.g. 129)10.
- Tumor susceptibility differences (e.g. C3H substrains)15.
Substrains are designated by specific nomenclature to help researchers identify their origin and history. Substrains are designated by appending a laboratory code (e.g. J for The Jackson Laboratory) to the end of a strain name without an additional forward slash. Laboratory codes are cumulative and are assigned by the Institute for Laboratory Animal Research (ILAR: dels.nas.edu/ilar/).
A substrain designation is added to a strain name if a colony has been separated from its progenitor strain for 20 or more generations, if genetic differences are detected, or for strains that are maintained completely independent of the progenitor strain. The websites of most mouse vendors provide information regarding the origin of substrains maintained at their facility, including the generation of separation from the founder strain, as recommended by the International Committee on Genetically Standardized Nomenclature for Mice.
There are many reasons JAX® Mice are the substrain of choice in the current environment of genome analysis and genetic manipulation:
- The genome of JAX® Mice strain C57BL/6J (Stock Number 000664) has been sequenced by the Mouse Genome Sequencing Consortium and it is, thus, the reference strain for mapping and comparison to the Human Genome16. For investigators relying on commercially available mouse sequence data, in addition to C57BL/6J, the reference strains A/J (Stock Number000646), DBA/2J (Stock Number 000671), 129X1/SvJ (Stock Number 000691), and 129S1/SvImJ (Stock Number 002448), were obtained and are available from The Jackson Laboratory17.
- JAX® Mice substrains were used for MIT SSLP data18 and for current SNP data efforts19.
- JAX® Mice substrains were selected for characterization by the Mouse Phenome Project (phenome.jax.org), whose goal is to establish a collection of phenotypic data on commonly used and genetically diverse mouse strains through a coordinated international effort20.
- JAX® Mice substrains were used to generate panels of consomic (chromosome substitution) strains for use as gene mapping tools21.
- The Complex Trait Consortium (www.complextrait.org) plans to use JAX® Mice substrains as progenitor strains for the generation of >500 recombinant inbred strains22.
- The Jackson Laboratory is the largest repository for genetically engineered and mutant mice. The majority of transgenes, spontaneous and induced mutations are maintained on JAX® Mice inbred strains, most commonly C57BL/6J. It is important to use matching inbred strains as experimental controls.
- Large amounts of reference data are available for JAX® Mice inbred strains. This includes strain-specific genetic (sequence and mapping data) and phenotypic information available through the JAX® Mice and Mouse Genome Informatics (www.informatics.jax.org) websites.
In large breeding facilities such as the production facility at The Jackson Laboratory, it is necessary to maintain pedigree traceability, maximize homogeneity within each strain, and generate large numbers of mice for distribution. The Jackson Laboratory employs systematic breeding and genetic quality control protocols to maximize homogeneity and minimize genetic drift.
The Jackson Laboratory utilizes three levels of expansion for each inbred strain (see Figure 1).
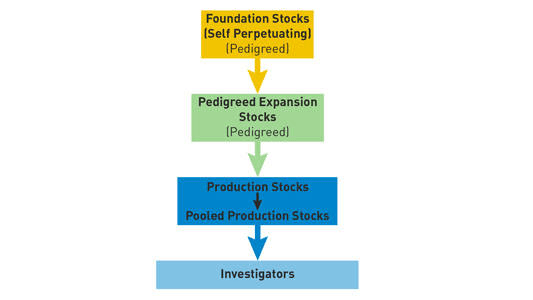
Figure 1. Relationship between foundation stocks, pedigreed expansion stocks, and production stocks at The Jackson Laboratory.
- Foundation stocks consist of select sibling breeding pairs of pedigreed inbred mice. They are used to produce additional breeding pairs and minimize generation number, thus limiting genetic drift. Foundation stocks maintained at The Jackson Laboratory are held separately from other stocks to ensure strict genetic and health controls. As part of our Genetic Quality Control program, every foundation stock animal is typed using single nucleotide polymorphic (SNP) markers.
- Pedigreed expansion stocks originate from foundation stocks and are propagated to generate sufficient numbers of mice to supply breeders to the production colonies. The number of generations arising from pedigreed expansion stocks is kept to a minimum.
- Production stocks consist of sibling breeding pairs and trios while pooledproduction stocks consist of non-sibling breeding pairs and trios. One generation of non-sibling mating is allowed for colony expansion. On rare occasions, a second generation of mating is permitted. Offspring from these stocks are distributed to researchers.
Using this system, colonies are expanded from a single ancestral pair to produce large numbers of genetically uniform mice in a minimum number of generations. This separation, plus cryopreservation of embryos from each strain, provides insurance against the loss of a strain in the event of fire, disease or other catastrophic event. The pedigree relationships in foundation stocks of all strains are controlled so that the breeding pairs in foundation stocks are traceable to a common ancestor in as few generations as possible.
References
(Authors in bold are Jackson Laboratory scientists)
- Davisson MT. The Future for Animal Models. Lab Anim 1999; 28:53-56.
- Paigen K. A miracle enough: the power of mice. Nat Med 1995; 1:215-220.
- Paigen K. Understanding the human condition: experimental strategies in Mammalian genetics.ILAR J 2002; 43:123-135.
- Zambrowicz BP and Sands AT. Knockouts model the 100 best-selling drugs-will they model the next 100?Nat Rev Drug Discov 2003; 2:38-51.
- Fox RR and Witham BA. Handbook of Genetically Standardized JAX® Mice. The Jackson Laboratory, Bar Harbor, ME (1997).
- Beck JA, et al. Genealogies of mouse inbred strains. Nat Genet 2000; 24:23-25.
- Specht CG and Schoepfer R. Deletion of the alpha-synuclein locus in a subpopulation of C57BL/6J inbred mice.BMC Neurosci 2001; 2:11.
- Sluyter F, Marican CC, and Crusio WE. Further phenotypical characterisation of two substrains of C57BL/6J inbred mice differing by a spontaneous single-gene mutation.Behav Brain Res 1999; 98:39-43.
- Bailey DW. Genetic drift: the problem and its possible solution by frozen?embryo storage.Ciba Found Symp 1977; 291-303.
- Simpson EM, et al. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice.Nat Genet 1997; 16:19-27.
- Threadgill DW, Yee D, Matin A, Nadeau JH, and Magnuson T. Genealogy of the 129 inbred strains: 129/SvJ is a contaminated inbred strain.Mamm Genome 1997; 8:390-393.
- Crabbe JC, Wahlsten D, and Dudek BC. Genetics of mouse behavior: interactions with laboratory environment.Science 1999; 284:1670-1672.
- Crawley JN, et al. Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies.Psychopharmacology (Berl) 1997; 132:107-124.
- Stiedl O, et al. Strain and substrain differences in context- and tone-dependent fear conditioning of inbred mice.Behav Brain Res 1999; 104:1-12.
- Glant TT, et al. Variations in susceptibility to proteoglycan-induced arthritis and spondylitis among C3H substrains of mice: evidence of genetically acquired resistance to autoimmune disease.Arthritis Rheum 2001; 44:682-692.
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome.Nature 2002; 420:520-562.
- Mural RJ, et al. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome.Science 2002; 296:1661-1671.
- Dietrich WF, et al. A comprehensive genetic map of the mouse genome.Nature 1996; 380:149-152.
- Lindblad-Toh K, et al. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse.Nat Genet 2000; 24:381-386.
- Paigen K and Eppig JT. A mouse phenome project. Mamm Genome 2000; 11:715-717.
- Nadeau JH, et al. Analysing complex genetic traits with chromosome substitution strains.Nat Genet 2000; 24:221-225.
- Vogel G. Genetics: Scientists dream of 1001 complex mice. Science 2003; 301:456-457.
