Summary: Pharmacogenomic test reports provide information about an individual's collection of genetic variants (or genotype) that cause them to metabolize or react to medicines in certain ways (phenotype). This resource provides information about common terminology and nomenclature used in pharmacogenomic test reports.
By: Clinical Education at JAX | April 2024
Pharmacogenomic test reports provide information about the impact of any variants identified on the function of the metabolizing enzyme. Often this information is provided in the context of dosing considerations for specific drugs.
Table 1: Common phenotypic results
Metabolizer status (Phenotype) | Functional Definition | Genetic Definition | Example Genotypes | Example Clinical Recommendations |
Ultrarapid metabolizer | Increased enzyme activity compared to rapid metabolizers | Two increased function alleles, or more than 2 normal function alleles | CYP2D6 *1/*1x2 (normal/two copies of normal allele) CYP2D6 *1/*45 (normal/two copies of normal allele) | Avoid codeine use because of potential for serious toxicity
|
Normal metabolizer | Fully functional enzyme activity | Combinations of normal function and decreased function alleles | CYP2D6 *1/*1 (normal/normal) CYP2D6 *1/*10 (normal/decreased)
| Use codeine label recommended age-specific or weight-specific dosing.
|
Intermediate metabolizer | Decreased enzyme activity (activity level between normal and poor metabolizer) | Combinations of normal function, decreased function, and/or decreased function alleles | CYP2D6 *1/*5 (normal/no function) CYP2D6 *4/*41 (decreased/decreased)
| Use codeine label recommended age-specific or weight-specific dosing.
|
Poor metabolizer | Little to no enzyme activity | Combination of no function alleles and/or decreased function alleles | CYP2D6 *5/*5 (no function/no function) CYP2D6 *4x2/*5 (two copies of no function allele/no function) | Avoid codeine use because of possibility of diminished analgesia. |
The genotype reported may be expressed in two different ways.
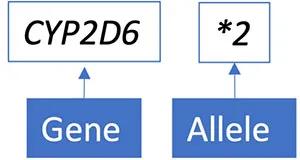
Most commonly, reports provide the genotypic results using star (*) nomenclature as an abbreviation for known alleles (different versions of a gene) containing different genomic variants.
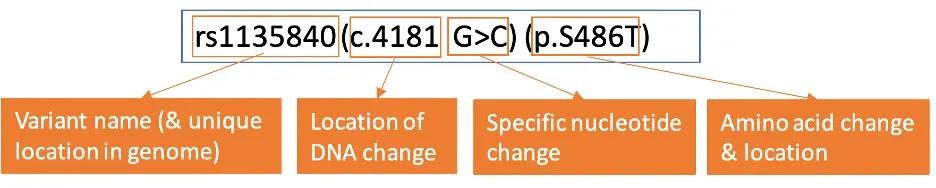
Some reports will also provide detailed information about the specific DNA variants present. In this example, the CYP2D6*2 allele contains two variants: c.2851C>T & c.4181G>C.

If not included on a report, the specific DNA variant(s) in an allele can also be identified through PharmVar and other resources. These genomic results provide data about the location of the variant and its impact on the resulting amino acid.
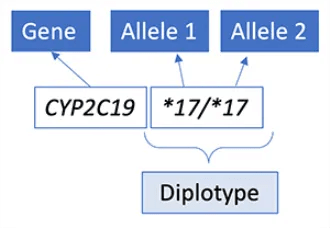
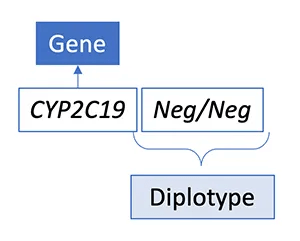
Results are commonly reported as a diplotype, which includes one maternal and one paternal allele that are both important to evaluate to predict clinical outcome.
In this example, CYP2C19*17/*17 denotes 2 increased function alleles (*17) inherited for the CYP2C19 gene, and this result predicts an ultrarapid metabolizer phenotype (see Table 1).
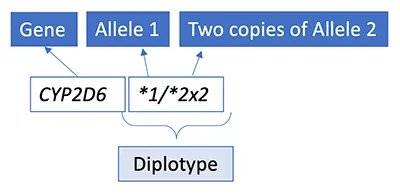
In some cases, patients have more than two copies of a gene, due to copy number changes (insertions, deletions, and duplications of segments of DNA are also collectively referred to as copy number variants).
Those alleles are denoted by an “xN” following the allele designation, for example, CYP2D6*1/*2x2.
Some reference laboratories will report an allele as “Neg” or Negative to indicate that no variants were detected by the assay.
A result of “Negative” for an allele may be interpreted as *1 or normal function.
Note: This result refers only to variants specifically tested by the lab. As with all genetic tests, ensure variants of interest were assessed by the lab before relying on a negative result.
Allele | One of two or more versions of a gene. An individual inherits two alleles for each gene, one from each parent. |
Copy Number
| Number of copies of a particular gene present in the genome of an individual. Insertions, deletions, and duplications of segments of DNA are also collectively referred to as copy number variants. |
Diplotype | The two alleles inherited by an individual for a particular gene. Includes one maternal and one paternal allele that are both important to evaluate to predict phenotype. |
Drug-metabolizing phenotype | Clinical presentation or observable characteristics of an individual with a particular genotype (e.g., poor metabolizer phenotype associated with CYP2C19*2/*2 genotype). |
Gene | Basic physical and functional unit of inheritance. |
Genotype | An individual's collection of genetic variants. |
Pharmacogenomics | Study of how genes affect the way a person responds to medications; pharmacogenomics may be used to determine ahead of time the best drug or dose for an individual patient. |
Phenotype | An individual’s observable traits. |
Single nucleotide variant or polymorphism | a single base pair addition, deletion, or substitution in a gene. A SNV or SNP may impact the function of a gene or its resulting protein, or it may have no effect at all. |
Star allele nomenclature | Common format used to represent variability of a specific gene; reported as gene symbol, (*) allele number/ (*) allele number (e.g.,CYP2C19*1/*2). |
Definitions provided by https://www.genome.gov/genetics-glossary
Exploring Pharmacogenomic Testing (CME | CNE). Learn how to determine whether pharmacogenomic testing is appropriate for the patient and practice applying test results to patient management.
Genomic Testing for the Healthy Individual (CME | CNE). Practice identifying patient motivations for genomic testing and assessing if a genomic test is a good fit.
Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, Whirl-Carrillo M, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19(2):215-223.
Crews KR, Monte AA, Huddart R, Caudle KE, Kharasch ED, Gaedigk A, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin Pharmacol Ther. 2021; Jan 2 doi: 10.1002/cpt.2149.
NHGRI, 2020. Talking glossary of genetic terms. Accessed February 20, 2021.
Pharmacogene Variation Consortium (PharmVar) at www.PharmVar.org (Gaedigk et al. 2018, CPT 103:399; Gaedigk et al. 2019, CPT 105:29)
Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics Knowledge for Personalized Medicine. Clin Pharmacol Ther. 2012; 92(4): 414-417.
Disclaimer
All information in this resource is provided for educational purposes only.