Alterations in the mouse microbiome have the potential to affect experimental results profoundly. Surprisingly, the microbiome of a mouse at JAX is quite stable, exhibiting only "micro-fluctuations" over time. Learn which microorganisms are found in 3 of the most commonly used mice in biomedical research: C57BL/6J, BALB/cJ, and NSG™.
Building Reproducibility through Microbiome Stability in JAX Mice
Blog Post | August 6, 2019
Impact of the Microbiome on Experimental Conclusions
Alterations in mouse health have the potential to affect experimental results profoundly. It’s easy to imagine that a mouse that is visibly ill (hunched, inactive, with poor body condition) may differ in phenotype from a visibly healthy mouse (bright, alert, and responsive).
But what about less visible changes, like alterations in the gut microbiome? For instance, a lab at Michigan State University experienced difficulties determining the effect of a particular drug on bone density, observing variable results in serial experiments despite controlling for a number of crucial variables. They later realized the inability to replicate their findings was due to differences in the microbiomes of their different cohorts of mice (Servik, 2016). Only after discovering this discrepancy were they able to assess the true impact of their drug on bone homeostasis.
Not every perturbation in the microbiome will have such impactful consequences. But identifying the microbial populations present in the mouse, in parallel with describing detailed breeding schemes, housing conditions, and caging configuration as suggested in the ARRIVE guidelines, may contribute a broader context for data interpretation and promote experimental reproducibility.
As a provider of reliable research models serving the biomedical community, JAX strives to provide mice of consistently high health status to investigators around the world. To support this mission, we set out to address a fundamental question among researchers: “how stable is the microbiome in individual JAX mouse strains over time?”
To begin to address this question, we designed a study characterizing the microbial milieu in three genetically distinct, commonly used mouse strains from a single, Maximum Barrier, Pathogen and Opportunistic-Free mouse production room at JAX. Overall, we found that the microbial communities do not vary significantly over five months for a given mouse strain in a given mouse room.
JAX Gut Microbiome Study Design
Fresh catch stool samples were collected from 3 randomly selected mice per cage. Samples included male and female mice starting at 3-5 weeks of age and were analyzed for five months. This resulted in a total of 2477 samples or roughly five samples/sex/timepoint/mouse strain. High density 16S V1-V3 rRNA sequencing was performed on each sample, and a total of 49.5 million amplicon sequences were analyzed for potential shifts in microbial community structure. Mouse strains analyzed
- C57BL/6J (Stock 000664), the most commonly used genetic background in microbiome research
- BALB/cJ (Stock 000651), a strain frequently used in comparative studies with C57BL/6J mice to assess the impact of immunological differences on host phenotype
- NSG™ or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (Stock 005557), a highly immunodeficient strain with applications for the study of host-specific factors and microbial colonization
Microbiomes of Individual Mouse Strains at JAX are Consistently Unique
The microbiomes of the three strains of JAX mice were identified using 16S ribosomal RNA sequencing (16S rRNA), an approach used to classify the bacterial communities present. Sequences were clustered when they shared 97% nucleotide identity, allowing for approximate genus-level comparisons across groups and resulting in a total of 347 operational taxonomic units (OTUs).
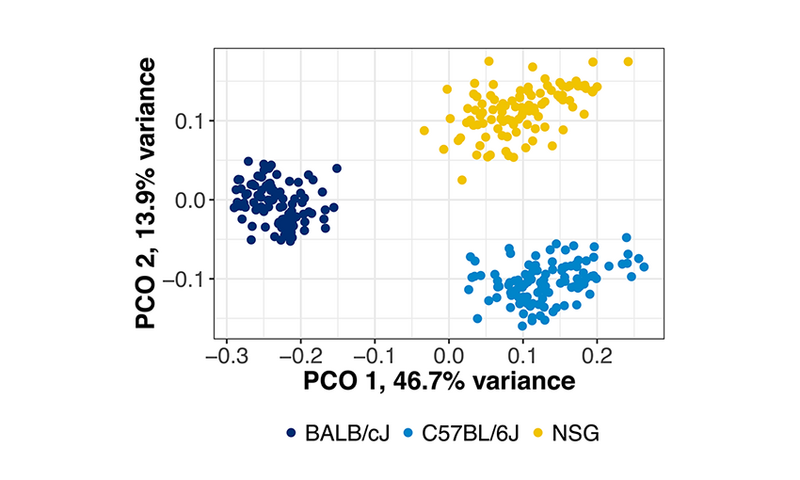
Relationships between microbial communities can be visually represented by Principal Coordinate Analysis (PCoA) plots. In these plots, points that are closer together represent samples with more similar microbial communities, while points located further from one another exhibit less similar microbial populations.
Interestingly, though each mouse strain exhibits a unique microbial structure, individual strains cluster together tightly within themselves:
What Organisms Form the Gut Microbial Universe of a JAX Mouse?
The 347 OTUs can be further collapsed into family-level taxonomic groups and reported as a percentage of relative abundance:

Broadly, the taxonomic families identified in these mouse strains are those known to maintain normal gut homeostasis and are commonly found in both healthy mice and healthy humans. All three mouse strains are primarily populated by Lachnospiraceae and Porphyromonadaceae, with BALB/cJ having the strongest representation of these families. Compared to BALB/cJ, C57BL/6J contain proportionally more family Erysipelotrichaceae and order Clostridiales. NSG™ have similar proportions of Erysipelotrichaceae and Clostridialesas to C57BL/6J, but also have an expansion of the Lactobacillaceae. Overall, while many individual microbes vary in abundance across the three strains (see PCoA plot above), the broader pattern of abundance at the family level is more similar.
Genetics and the relative immune function of these mice likely play an active role in determining the microbial universe of each strain. Certainly, many researchers worldwide are working to assess the impact of these variables on host bacterial communities.
Microbiomes of Individual Mouse Strains at JAX are Highly Stable Over Time
Importantly, when the same PCoA data is visualized at 2-week time intervals, no obvious skewing of microbial populations is apparent:

Indeed, time points frequently overlap each other with minimal shifts in overall bacterial community structure. This is a significant finding considering that feed, water, bedding, and caging were refilled and changed in the mouse room several times throughout the five months, and underscores JAX's commitment to optimal husbandry practices.
How is Microbiome Stability Achieved at JAX?
Together, these data suggest that within a particular strain at JAX, microbial communities vary quite little over time.
It’s noteworthy that these experiments were conducted in a mouse production room that supplies mice to laboratories around the world. JAX production rooms are highly controlled to restrict the introduction of pathogens and opportunists through careful sterilization, including autoclaving of feed, drinking water, bedding, individually ventilated microisolator caging, supplies, and equipment. Animal technicians adhere to a rigorous room entry procedure which includes full gowning with multiple forms of personal protective equipment, including face and head coverings and finishing up with an air-shower entry. Health monitoring is performed every six weeks on animals in the room, and diagnostic testing of feed, water, housing surfaces, and equipment is completed to validate these processes.
Through rigorous housing and husbandry processes, including tightly controlled barriers and an animal health monitoring program, it is possible to maintain microbiome stability in laboratory mice as a foundation for experimental reproducibility. At JAX, we are dedicated to these processes and maintaining our facilities at the highest standards to best support your research needs.
What will you do when your JAX mice arrive to ensure stability in their microbiome?
References
Servik K., 2016. Of mice and microbes. Science. 353(6301):741-3. PMID: 27540148. [DOI: 10.1126/science.353.6301.741]
