Female and male C57BLKS/J mice with the leptin receptor mutation (BKS-db; BKS.Cg-Dock7m +/+ Leprdb/J) develop type 2 diabetes with phenotypes that include:

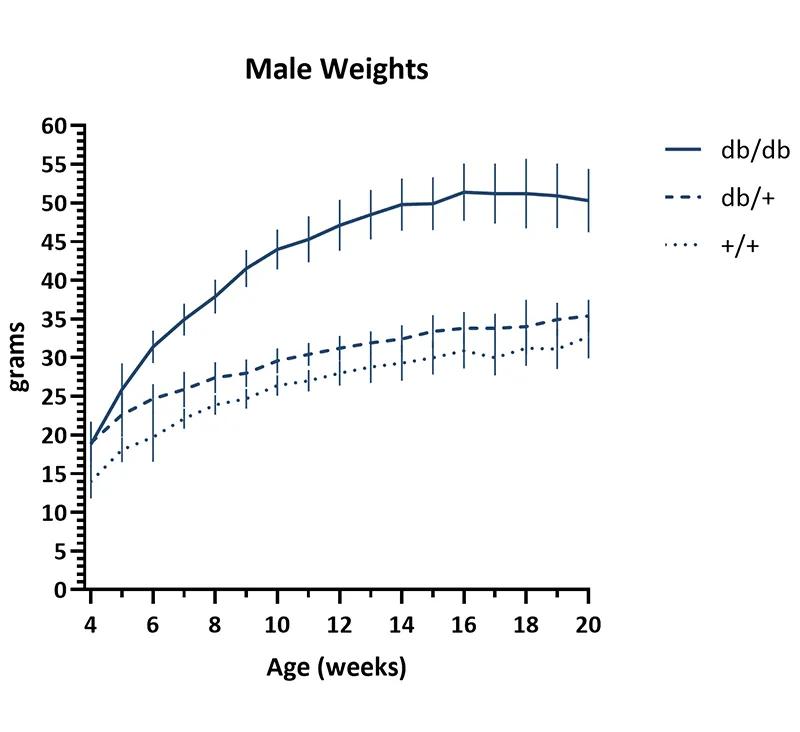
Figure 1 Body weight growth curve. Weekly body weights of female (left) and male (right) mice. Cohorts included 60-70 db/db homozygous, 35 db/+ heterozygous, and 32-33 wildtype (+/+) mice per sex. Mice were fed a diet containing 6% fat (LabDiet® 5K52 formulation). Values represent mean and one standard deviation.


Figure 2 Non-fasted glucose. Blood glucose levels in female and male mice were measured every other week. Submental blood glucose measurements were obtained from the same cohorts described in Figure 1 using a OneTouch Ultra 2 or UltraMini hand-held glucometer that was validated with a control glucose solution on each day of use. Values represent mean and one standard deviation.


Figure 3 Glucose tolerance test. Following a 16 hour fast, serum was collected from submental blood for an initial glucose reading (“Before”) and mice were then administered glucose by IP injection at 2g/kg body weight. Serum glucose was measured after 120 minutes (“After”). Values represent mean and one standard deviation of 10-15 mice per genotype and age. Glucose was measured with a Beckman Coulter DxC 700 AU chemistry analyzer.

Figure 4 Body Composition. Mice were analyzed using a Lunar PIXImus DEXA scanner. Calculations of body composition exclude the head. Values represent mean and one standard deviation of 10 non-fasted db/db and 4-5 db/+ & +/+ mice per sex and age. Results were analyzed separately by age and sex using two-way ANOVA with Sidak's multiple comparisons test to identify values that differed significantly between genotypes, using GraphPad Prism version 10.0 for Windows (GraphPad Software). An * indicates that the db/db result was significantly different from both control genotypes at that age (P < 0.0001).








Figure 5 Serum chemistry and hormones. All values were measured from serum collected from submental blood except HbA1c, which was measured from submental whole blood. Values represent mean and one standard deviation of the same 20 non-fasted db/db mice and 10 db/+ & +/+ mice per age, studied longitudinally. Results were obtained using a Beckman Coulter DxC 700 AU chemistry analyzer or Meso Scale SQ120 analyzer (insulin & leptin). Results were analyzed separately by parameter using a repeated measures mixed-effects model with Sidak's multiple comparisons test. An * indicates that the db/db result was significantly different from both control genotypes at that age (P < 0.05).
Table 1 Distribution of db/db blood glucose values. The blood glucose values of db/db females and males shown in Figure 2 were grouped into ranges. Rows indicate the percentage of mice in the indicated ranges.
| Glucose Range (mg/dL) | 4 weeks | 6 weeks | 8 weeks | 10 weeks | 12 weeks | 14 weeks | 16 weeks | 18 weeks | 20 weeks | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | |
| < 250 | 63% | 60% | 21% | 29% | 19% | 16% | 5% | 2% | 0% | 3% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| 250 - 349 | 18% | 30% | 40% | 37% | 21% | 29% | 24% | 36% | 7% | 15% | 5% | 12% | 2% | 0% | 0% | 0% | 6% | 0% |
| 350 - 449 | 20% | 8% | 29% | 16% | 26% | 39% | 42% | 38% | 29% | 40% | 29% | 49% | 13% | 35% | 19% | 15% | 13% | 0% |
| 450 - 599 | 0% | 3% | 10% | 16% | 33% | 16% | 29% | 24% | 64% | 43% | 58% | 39% | 73% | 57% | 73% | 67% | 52% | 67% |
| > 600 | 0% | 0% | 0% | 2% | 0% | 0% | 0% | 0% | 0% | 0% | 7% | 0% | 13% | 8% | 8% | 19% | 29% | 33% |