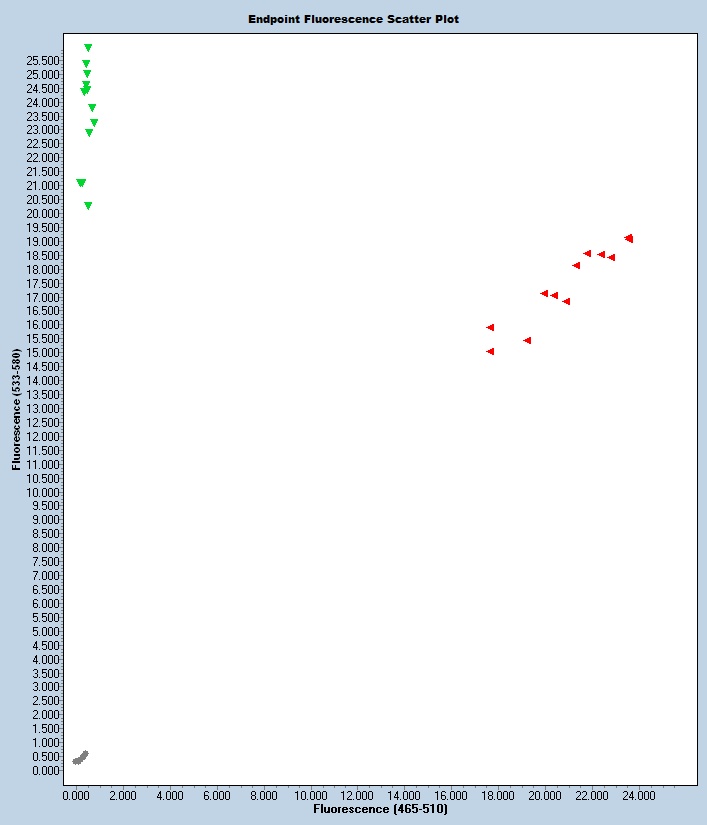
Notes
Taqman qPCR protocols are run on a real time PCR instrument. Use an appropriate instrument specific Fluorophore/Quencher combination.
Expected Results
Mutant= 84 bp
Wild Type = 94 bp
>chr11:33518210-33518303 94bp TGGGTATTGCTTTTATAATTTCTCG ATTGCATCCTCTGCTCCATT |
Sequence
Wt Sequence (deletions in lower case; insertions with carrots ^g^ (g insertion):
TGGGTATTGCTTTTATAATTTCTCGAGTTTTGTTTCCAACCCAC^A^taatgtagatacaccggctgcggagtctgaaatggagcagaggatgcaattggtggttagatggaggagagcttgtgtttgtgtttctatgtaagtctttctgttttttttttcttccttctcaggtgacccagttcggatagcactgactctggacatcgccagcatctctagcatctcagagagtaacatggtaaccagtgccacccccaccccttcccccgctccctctccttgctttcaattgtaatgtcctttcaagtaaatcagaaacagagtggcagatgccagagttctgtcttgtaagtggagacagtagatgtttataggttttctgggtatcaggaagggaacagactcaaaggccaggaagagagaaagtagaaaaggaagtgaccgattgactgaggaataaaaacatgtccacacgGTCTTTCAGTCGAGGGACAGCTGATAAAACTTACAATGT
This mutation is a 430 bp deletion beginning at Chromosome 11 position 33,517,830 bp and ending after 33,518,259 bp with a signle bp A insertion at the junction(GRCm39/mm39).
JAX Protocol
Protocol Primers
| Primer | 5' Label | Sequence 5' → 3' | 3' Label | Primer Type | Reaction | Note |
|---|---|---|---|---|---|---|
| 65043 | TGG GTA TTG CTT TTA TAA TTT CTC G | Common | A | |||
| 65044 | ATT GCA TCC TCT GCT CCA TT | Wild type Reverse | A | |||
| 65045 | ACA TTG TAA GTT TTA TCA GCT GTC C | Mutant Reverse | A | |||
| 65046 | Fluorophore-1 | ATG TAG ATA CAC CGG CTG CG | Quencher-1 | WT Probe | ||
| 65047 | Fluorophore-2 | TTC CAA CCC ACA GTC TTT CAG TC | Quencher-2 | MUT Probe |
Reaction A
| Component | Final Concentration |
|---|---|
| Kapa Probe Fast QPCR | 1.00 X |
| ddH2O | |
| 65043 | 0.40 uM |
| 65044 | 0.40 uM |
| 65045 | 0.40 uM |
| Wt Probe | 0.15 uM |
| Mutant Probe | 0.15 uM |
| DNA |
Cycling
| Step | Temp °C | Time | Note |
|---|---|---|---|
| 1 | 95.0 | -- | |
| 2 | 95.0 | -- | |
| 3 | 60.0 | -- | |
| 4 | -- | repeat steps 2-3 for 40 cycles | |
| 5 | 40.0 | -- | Forever |