What is a DREADD and how can I use it?
DREADDS - Designer Receptors Exclusively Activated by Designer Drugs - are engineered G-protein coupled receptors that are unresponsive to endogenous ligands, but can be activated by nanomolar concentrations of pharmacologically inert and metabolically stable small molecules. Where they are expressed, DREADDS allow precise spatiotemporal control of GPCR signaling. In 2007, Dr. Bryan L. Roth at the University of North Carolina, Chapel Hill developed DREADDS derived from muscarinic receptors that were no longer activated by their endogenous ligand acetylcholine but instead were activated by the small molecule clozapine-N-oxide (CNO) (Armbruster et al. 2007). Ultimately, Dr. Roth’s team designed DREADDS that enabled non-invasive control of neuron signaling through the Gq (hM3Dq), Gi (hM4Di) and Gs (Gs-D) G-protein coupled signaling pathways. Previously, DREADD protein expression has been introduced and controlled in vivo using tetracycline-dependent transgenes, or cre-dependent or cre-independent viral vectors. Now, Dr. Roth’s team in collaboration with Dr. Ute Hochgeschwender’s group from Duke University have developed multiple cre recombinase-conditional strains that conditionally express either an activating (hM3Dq) or an inhibiting (hM4Di) DREADD in cells where cre is active. These mice can be combined with the already extensive collection of cre-expressing strains to study Gq and Gi-mediated signaling pathways in a wide variety of neurons and tissues.
First Step: Create Your DREADD Targeting Vector
Dr.'s Roth and Hochgeschwender designed their cre-conditional DREADD constructs based on the targeting vector that the Allen Institute for Brain Science used to create Ai9 cre reporter mice (e.g. B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (007909)). In their vectors, the North Carolina teams replaced the tdTomato reporter cassette with wither HA-tagged hM3Dq- or hM4Di-DREADD coding sequences linked to a downstream fluorescence reporter cassette via a P2A ribosomal entry site. The bi-cistronic DREADD/reporter sequence is preceded upstream by a CAG promoter and a loxP-flanked STOP codon. The modifed Ai targeting vector, Rosa-CAG-LSL-HA-DREADD-P2A-mCitrine, allows for robust transgene expression after cre-mediated excision of the upstream STOP sequence. The HA-tag allows for detailed spatial localization of the DREADD receptors after cre-excision via immunohistochemical staining. To test whether this new vector functioned properly, the Roth and Hochgeschwender team introduced a cre-expressing plasmid into ES cells that carried the Rosa-CAG-LSL-HA-DREADD-P2A-mCitrine construct. As expected, only ES cells carrying the construct and transfected with the cre vector fluoresced green.
Second Step: Create Your Mouse Model
Mice heterozygous for Rosa-CAG-LSL-HA-DREADD-P2A-mCitrine allele are viable and fertile. For DREADD analysis, the mice were subsequently mated to various Cre driver strains available at The Jackson Laboratory. The cre strains used and the resulting data are summarized below:
- B6.C-Tg(CMV-cre)1Cgn/J (006054) – Deletion of loxP flanked genes in all tissues. LSL-hM4Di X CMV-Cre and LSL-hM3Dq X CMV-Cre resulted in wide-spread DREADD expression in peripheral organs.
- B6.Cg-Tg(Camk2a-cre)T29-1Stl/J (005359) – Deletion of loxP flanked genes in tissues expressing Camk2a, primarily in the forebrain and specifically the CA1 pyramidal cell layer in the hippocampus. Both hM4Di and hM3Dq expression was consistent with the characterized expression pattern of the Camk2a-cre line.
- B6.129S2-Emx1tm1(cre)Krj/J (005628) – Deletion of loxP flanked genes in tissues expressing Emx1 - localized to neurons of the neocortex and hippocampus, including glial cells of the pallium. Both hM4Di and hM3Dq expression was consistent with the characterized expression pattern of the Emx1-Cre line, mimicking the pattern of expression of the endogenous Emx1 gene.
A.
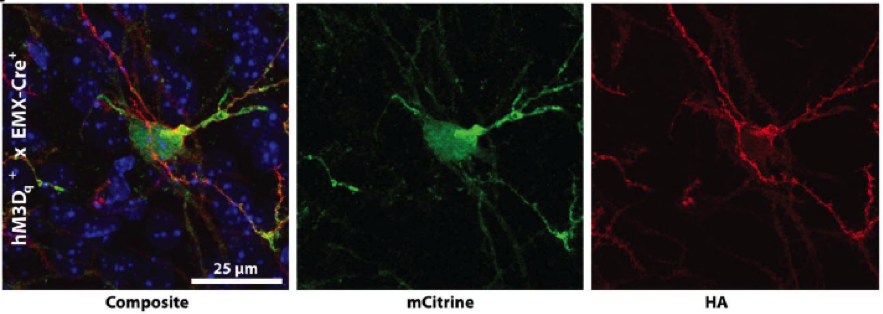
B.
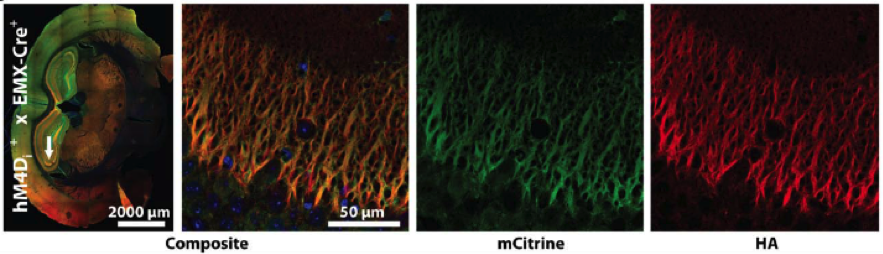
Figure. Conditional DREADD expression in targeted brain regions of Emx1-Cre mice. (A) Confocal images of neurons expressing HA-tagged hM3Dq DREADD at the membrane of mCitrine-positive neurons after Cre recombination. (B) hM4Di-Emx1-Cre mice with expression in mossy fiber projections to CA3 from the dentate gyrus after Cre recombination.
Third Step: Test the Functionality of Your Newly Created Conditional DREADD Mouse Model
Emx1-cre and LSL-hM3Dq double-positive offspring from the from the LSL-hM3Dq X Emx1-Cre cross were injected with CNO and were assessed behaviorally in the open field assay. Because Emx1 is expressed in excitatory cortical glutamatergic projection neurons, motor activity in the open field setting was increased in the CNO-treated mice compared with both vehicle-treated Emx1-cre/LSL-hM3Dq double-positive mice and CNO-treated LSL-hM3Dq-negative littermate controls. In vivo functionality was also assessed in LSL-hM4Di X Camk2a-Cre progeny to evaluate the effects of CNO-induced neuron silencing capabilities. Previous studies utilizing hM4Di-DREADDs to suppress hippocampal neuronal activity resulted in impaired consolidation of contextual memory in a contextual conditioned fear paradigm (Zhu et al., 2014). Consistent with these findings, LSL-hM4Di X Camk2a-Cre progeny expressing hM4Di in Camk2a showed effects in this assay.
With the vast selection of Cre-driver lines that are available, these new DREADD lines can allow for Cre-mediated DREADD expression that is both pathway and cell type-specific. They represent a novel platform that can be applied to many areas of research.
The conditional-ready hM3Dq and hM4Di DREADD lines are readily available now from The Jackson Laboratory:
- B6N.129-Gt(ROSA)26Sortm1(CAG-CHRM4*,-mCitrine)Ute/J (026219)
- B6N;129-Gt(ROSA)26Sortm2(CAG-CHRM3*,-mCitrine)Ute/J (026220)
Reference:
Zhu H, Aryal DK, Olsen RHJ, Urban DJ, Swearingen A, Forbes S, Roth BL, Hochgeschwender U. 2016. Cre-dependent DREADD (Designer receptors exclusively activated by designer drugs) mice. Genesis. Epub. PMID: 27194399
Additional references:
Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. 2007. Evolving the lock to fit the key to create a family of g protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A 104:5163–5168. PMID: 17360345
Zhu H, Pleil KE, Urban DJ, Moy SS, Kash TL, Roth BL. 2014. Chemogenetic inactivation of ventral hippocampal glutamatergic neurons disrupts consolidation of contextual fear memory. Neuropsychopharmacology 39:1880-1892. PMID: 24525710