Mutations in SARS-CoV-2 are generating viral variants that change the properties of the virus and affect the response to the COVID-19 pandemic. While the implications are still not entirely clear, preliminary studies indicate that, at the very least, a mutation common to three new variants of concern appears to increase transmission efficiency. The emergence and potential spread of the variants underscores the necessity to roll out vaccinations as quickly as possible, as well as expand genomic surveillance efforts to detect variants as they arise and travel around the globe.
Graphic by Jane Cha, The Jackson Laboratory.
The SARS-CoV-2 virus, which causes COVID-19, stayed relatively stable through last fall. Now all of a sudden, many variants are being identified that appear to change how the virus behaves. How does a virus change, and why are these variants emerging so quickly now?
To put it simply, viruses are designed to reproduce quickly, but not with 100% accuracy all the time. Viruses that have RNA genomes, including coronaviruses, usually mutate at a particularly high rate, because the proteins that copy RNA are error prone. Indeed, it was a mutation (or mutations) in the “spike” protein of SARS-CoV-2, likely in late 2019, that allowed it to emerge from its animal host(s) and efficiently infect humans. SARS-CoV-2 doesn’t mutate as fast as many other coronaviruses, however, because of its “spell-check” mechanism, an additional protein that corrects major copying mistakes. So during the first wave of infections last spring, many minor variants were tracked but nothing of particular concern emerged.
With the huge surge in infections worldwide since October, however, the virus has had millions of opportunities to mutate and evolve. Most of the changes have little or no effect, and they can even hamper transmissibility, meaning those variants will quickly disappear. But of all of these changes, a select few of them will actually enhance virus spread (e.g., in B.1.1.7, the variant first identified in England). If the variant is significantly more infectious, it can out-compete the other ones and become the most common found in humans over time.
What new variants are people tracking?
Note: The COVID-19 pandemic has grown and evolved so rapidly, even scientists are struggling to keep up. Therefore, the nomenclature of SARS-CoV-2 variants—what they are called when they arise—is still, in the words of the prestigious scientific journal Nature, “a bloody mess.” Therefore, while the following descriptions use currently common designations, they may change over time.
Graphic by Jane Cha, The Jackson Laboratory.
The variant of SARS-CoV-2 that originally arose in China and spread around the world is called D614G. It held sway throughout the spring and summer, and though minor changes enabled the tracking of different transmission routes, they all functioned in the same ways. Pharmaceutical companies, therefore, used D614G’s spike protein sequence to develop the vaccines that are in use today.
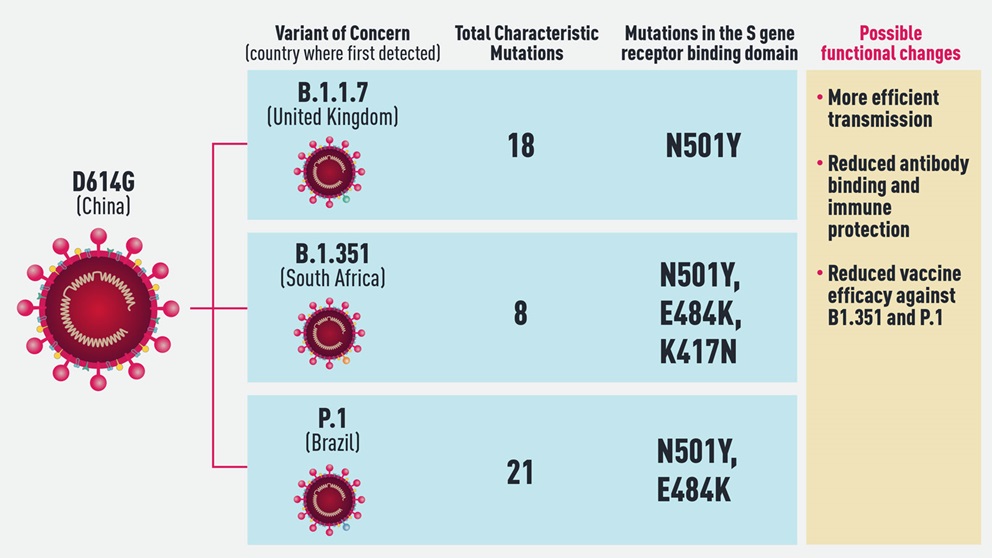
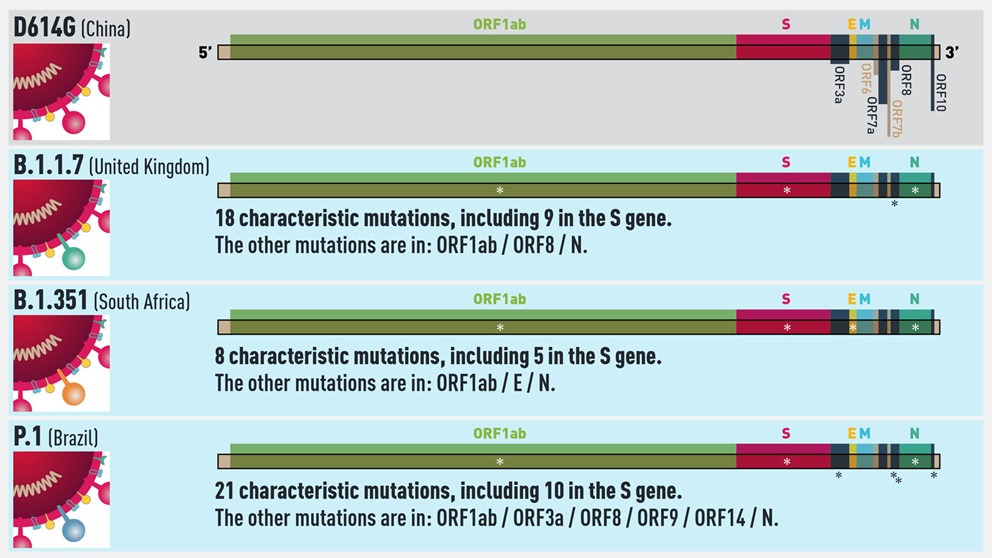
The emerging variant that has received the most attention and concern to date is known as B.1.1.7. First found in the U.K., B.1.1.7 is more efficiently transmitted than other variants identified so far. It carries 18 lineage-defining mutations, including one in the spike (S) gene of the virus known as N501Y that affects binding to the human ACE2 receptor. Data is still being compiled and analyzed, but early findings suggest it is also somewhat more virulent than D614G, causing severe disease in a higher percentage of patients. There is also good news, however, in that the first studies to come out indicate that the current vaccines fully protect against B.1.1.7, further increasing the pressure to vaccinate as many people as quickly as possible.
Less is known about the other two major variants on the radar: B.1.351, which first emerged in South Africa, and P1, which arose in Brazil and may be the cause of a highly concerning second wave of infection in the city of Manaus. B.1.351 has the same N501Y S gene mutation as the B.1.1.7 variant, and two more as well in key sites of the receptor binding domain of the spike protein. Furthermore, it may not be well recognized by the immune systems of people previously infected with D614G. Studies already show that vaccine reactivity is not as strong to B.1.351, but it should still be strong enough to be effective. More research is ongoing regarding both immune and vaccine protection against the variant.
Interestingly, in addition to many other lineage defining mutations, P.1 also has the N501Y mutation. Given that it spontaneously arose in three variants under investigation for enhanced transmissibility, N501Y likely provides a competitive advantage for the variants in which it’s found. Also of concern regarding P.1 is its emergence in Manaus, a region that previously underwent a massive surge of COVID-19—with infection rates of up to 75%, according to some estimates—implying that it may be able to re-infect people originally infected with a different variant. P.1 was not identified prior to December 2020, however, so its properties and potential for immune evasion are just now being studied.
How do scientists find new variants of the virus around the world?
To detect new variants of the virus, researchers isolate and sequence the genomes of as many patient samples as is feasible. It’s called genomic surveillance, and it makes it possible to track transmission patters, incidence of different variants, and so on. Unfortunately, genomic surveillance is somewhat expensive and takes time, so it’s often under-utilized, and variants can evade detection for crucial days, weeks and even months.
As a rule of thumb, sequencing about five percent of patient samples is considered necessary for good surveillance. At this time the U.K., in the midst of a severe outbreak driven by the B.1.1.7 variant, is sequencing approximately 10 percent of its cases. On the other hand, the U.S., with the largest patient population in the world, is only sequencing 0.3 percent of cases. And some countries have no genomic surveillance programs at all. Moving forward, it is hugely important to expand genomic surveillance, track the evolution and spread of the virus, and detect variants as they emerge. Even though variants such as B.1.1.7 are causing serious surges, we have been lucky in a way that nothing worse has emerged. If something does, it’s imperative to limit its spread and contain the damage as much—and as soon—as possible.