For in-depth product & services help, ask our
Technical Information Scientists
Stock No: 030890
Protocol 19800: Probe Assay - Il2rg<tm1Wjl>-PROBE
Version 4.0
Notes
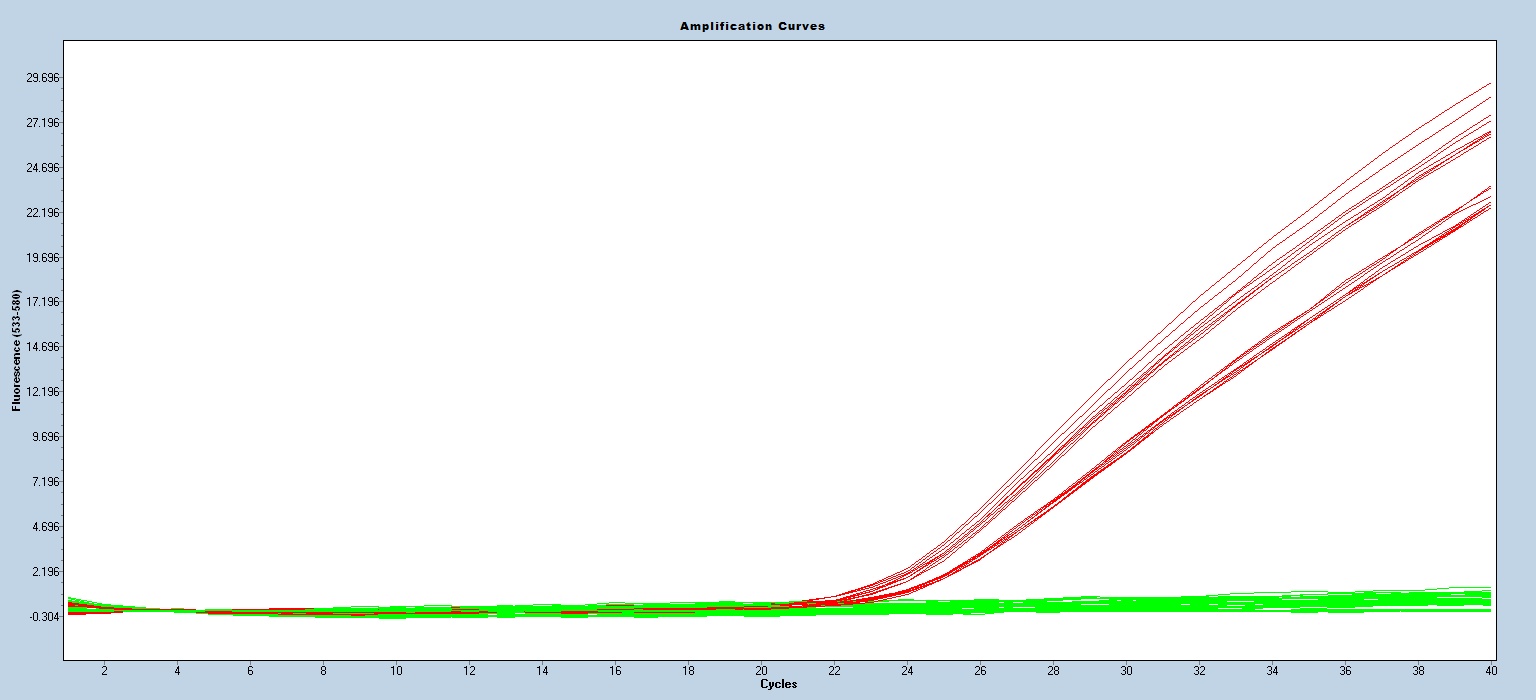
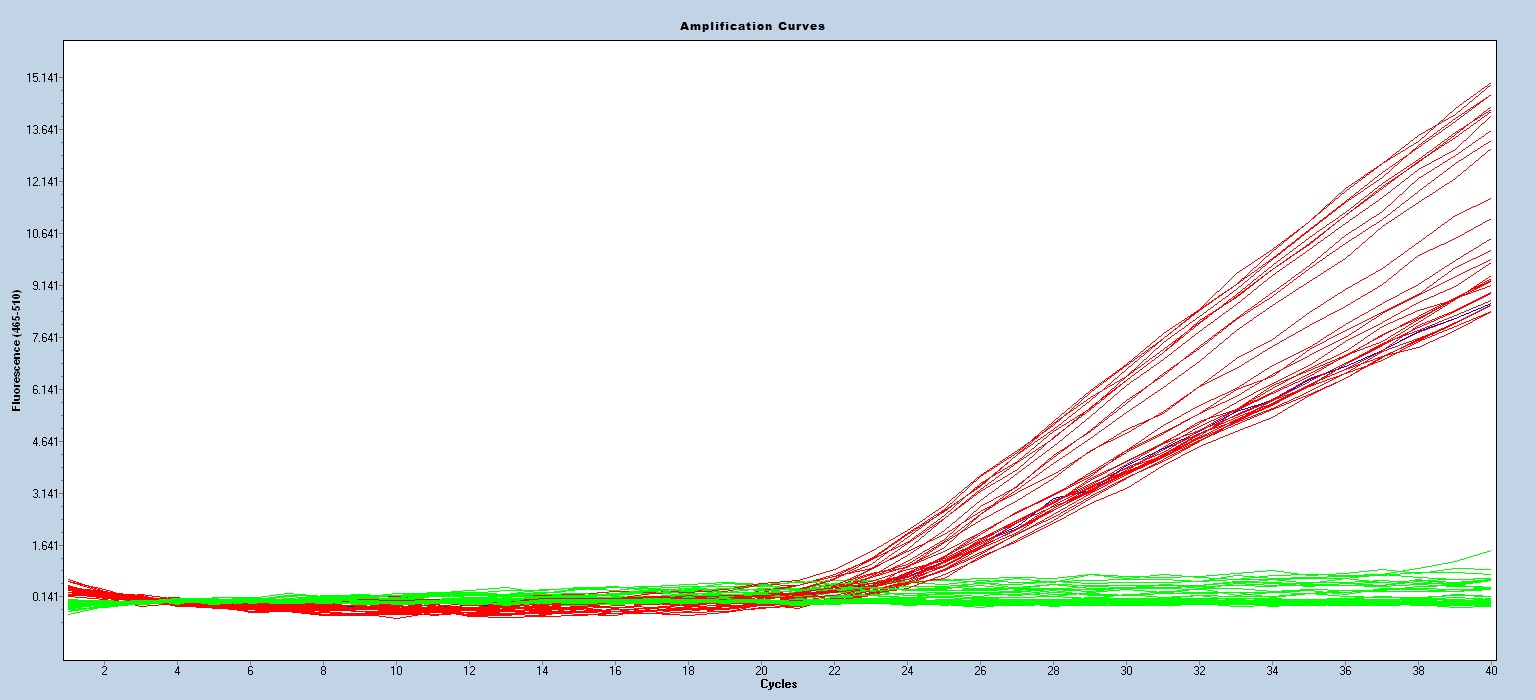
Taqman qPCR protocols are run on a real time PCR instrument. Use an appropriate instrument specific Fluorophore/Quencher combination. The transgene genotype is determined by comparing ΔCt values of each unknown sample against known homozygous and hemizygous controls, using appropriate endogenous references.
The genotyping protocol(s) presented here have been optimized for reagents and conditions used by The Jackson Laboratory (JAX).
To genotype animals, JAX recommends researchers validate the assay independently upon receipt of animals into their facility.
Reaction cycling temperature and times may require additional optimization based on the specific genotyping reagents used.
Expected Results
Mut-130bp
WT-94bp
Fam=Mut
Hex=wt
Cy5=IPC
Sequence
Mutant sequence, including junction:
TTCAGGAACCCTACCAGTTTCTCATGGGATGCATTGTCAGTTCAGACCAGATGAGGCTAGCT
AATGGGCATATGCATGCCCATGTTTGGCCCATCATTCTTTTGCCTTGTAACCCTTCTCTAGGT
ACAAGGTATCTGATAATAATACATTCCAGGAGTGCAGTCACTATTTGTTCTCCAAAGAGATT
ACTTCTGGCTGTCAGATACAAAAAGAAGATATC(aagcttatcgataccgtcgacctcgagggcccctgcag
gtcaattctaccgggtaggggaggcgcttttcccaaggcagtctggagcatgcgctttagcagccccgctgggcacttggcgct
aca)
WT Il2rg, including junction:
ttcaggaaccctaccagtttctcatgggatgcattgtcagttcagaccagatgaggctagct
aatgggcatatgcatgcccatgtttggcccatcattcttttgccttgtaacccttctctagGT
ACAAGGTATCTGATAATAATACATTCCAGGAGTGCAGTCACTATTTGTTCTCCAAAGAGATT
ACTTCTGGCTGTCAGATACAAAAAGAAGATATC(cagctctaccagacatttgttgtccagctccaggacccc
cagaaaccccagaggcgagctgtacagaagctaaacctacagaatcttggtaatcgggaaagaagtagccaagagagcagg
gagcttaaagacactggagtttatagattg)
JAX Protocol
Protocol Primers
| Primer | 5' Label | Sequence 5' → 3' | 3' Label | Primer Type | Reaction | Note |
|---|---|---|---|---|---|---|
| 19475 | AAG AGA TTA CTT CTG GCT GTC AG | Common | A | |||
| 19476 | Fluorophore-1 | AAA GCG CCT CCC CTA CCC G | Quencher-1 | MUT Probe | PGK | |
| 19477 | CTC TGG GGT TTC TGG GG | Wild type Reverse | A | |||
| 19478 | Fluorophore-2 | TGG AGC TGG ACA ACA AAT GTC TGG T | Quencher-2 | WT Probe | ||
| oIMR3914 | ATG CTC CAG ACT GCC TTG | Mutant Reverse | A |
Reaction A
| Component | Final Concentration |
|---|---|
| Kapa Probe Fast QPCR | 1.00 X |
| ddH2O | |
| 19475 | 0.40 uM |
| 19477 | 0.40 uM |
| oIMR3914 | 0.40 uM |
| Wt Probe | 0.15 uM |
| Mutant Probe | 0.15 uM |
| DNA |
Cycling
| Step | Temp °C | Time | Note |
|---|---|---|---|
| 1 | 95.0 | -- | |
| 2 | 95.0 | -- | |
| 3 | 60.0 | -- | |
| 4 | -- | repeat steps 2-3 for 40 cycles | |
| 5 | 40.0 | -- | Forever |
JAX uses a very high speed Taq (~1000 bp/sec), use cycling times recommended for your reagents.
Strains Using This Protocol
| Stock Number | Strain Name |
|---|---|
| 032088 | B6.129S-Rag1tm1Mom Cd47tm1Fpl Il2rgtm1Wjl/SzJ |
| 025730 | B6.129S-Rag2tm1Fwa Cd47tm1Fpl Il2rgtm1Wjl/J |
| 003174 | B6.129S4-Il2rgtm1Wjl/J |
| 039001 | B6.Cg-Rag2tm1Fwa Sgshmps3a Cd47tm1Fpl Il2rgtm1Wjl/JnolJ |
| 003169 | C.129S4-Il2rgtm1Wjl/J |
| 030547 | NOD.Cg-B2mtm1Unc Prkdcscid H2dlAb1-Ea Il2rgtm1Wjl/SzJ |
| 010636 | NOD.Cg-B2mtm1Unc Prkdcscid Il2rgtm1Wjl/SzJ |
| 038279 | NOD.Cg-Capn3em5Lutzy Prkdcscid Il2rgtm1Wjl/VitaJ |
| 031900 | NOD.Cg-Cmahem1Guru Prkdcscid Il2rgtm1Wjl/GuruJ |
| 028615 | NOD.Cg-Fcgrttm1Dcr Prkdcscid Il2rgtm1Wjl Tg(FCGRT)32Dcr/J |
| 035844 | NOD.Cg-Flt3em1Akp Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav Tg(HLA-A/H2-D/B2M)1Dvs/J |
| 035843 | NOD.Cg-Flt3em1Akp Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/J |
| 035842 | NOD.Cg-Flt3em1Akp Prkdcscid Il2rgtm1Wjl/J |
| 033367 | NOD.Cg-Flt3em2Mvw Prkdcscid Il2rgtm1Wjl Tg(FLT3LG)7Sz/SzJ |
| 026263 | NOD.Cg-Foxn1em1Dvs Prkdcscid Il2rgtm1Wjl/J |
| 029819 | NOD.Cg-Gcgem1Dvs Prkdcscid Il2rgtm1Wjl/DvsJ |
| 035959 | NOD.Cg-Gt(ROSA)26Sorem27(KRT18-ACE2)Mvw Prkdcscid Il2rgtm1Wjl/MvwJ |
| 038123 | NOD.Cg-Gt(ROSA)26Sorem28(KRT18-ACE2)Mvw Prkdcscid Il2rgtm1Wjl/MvwJ |
| 036151 | NOD.Cg-Gt(ROSA)26Sorem5Mvw Prkdcscid Il2rgtm1Wjl/MvwJ |
| 030511 | NOD.Cg-Hc1 Prkdcscid Il2rgtm1Wjl/SzJ |
| 014553 | NOD.Cg-Hgftm1.1(HGF)Aveo Prkdcscid Il2rgtm1Wjl/J |
| 035868 | NOD.Cg-Iduaem1Osp Prkdcscid Il2rgtm1Wjl/J |
| 035007 | NOD.Cg-Iduatm1Clk Prkdcscid Il2rgtm1Wjl/J |
| 026622 | NOD.Cg-KitW-41J Tyr + Prkdcscid Il2rgtm1Wjl/ThomJ |
| 026497 | NOD.Cg-KitW-41J Prkdcscid Il2rgtm1Wjl/WaskJ |
| 009617 | NOD.Cg-Mcph1Tg(HLA-A2.1)1Enge Prkdcscid Il2rgtm1Wjl/SzJ |
| 032474 | NOD.Cg-Prkdcscid Abcd1em2Lutzy Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ |
| 027905 | NOD.Cg-Prkdcscid Cd274tm1Shr Il2rgtm1Wjl/J |
| 030610 | NOD.Cg-Prkdcscid Cybbem1Hmal Il2rgtm1Wjl/HmalJ |
| 031566 | NOD.Cg-Prkdcscid H2-Ab1b-tm1Doi Il2rgtm1Wjl Tg(H2-Ea-HLA-DRB1*0401*)1Dv/SzJ |
| 026936 | NOD.Cg-Prkdcscid H2-Ab1b-tm1Doi Il2rgtm1Wjl Tg(HLA-DQA1,HLA-DQB1)1Dv Tg(INS*)172Dvs/DvsJ |
| 026561 | NOD.Cg-Prkdcscid H2-Ab1b-tm1Doi Il2rgtm1Wjl Tg(HLA-DQA1,HLA-DQB1)1Dv/SzJ |
| 021885 | NOD.Cg-Prkdcscid H2-Ab1b-tm1Doi Il2rgtm1Wjl/SzJ |
| 025216 | NOD.Cg-Prkdcscid H2-K1b-tm1Bpe H2-Ab1g7-em1Mvw H2-D1b-tm1Bpe Il2rgtm1Wjl/SzJ |
| 023848 | NOD.Cg-Prkdcscid H2-K1b-tm1Bpe H2-D1b-tm1Bpe Il2rgtm1Wjl/SzJ |
| 032927 | NOD.Cg-Prkdcscid H2-K1d-em1Dvs H2-Ab1g7-em1Dvs H2-D1b-em5Dvs Il2rgtm1Wjl/SzJ |
| 026222 | NOD.Cg-Prkdcscid Hprt1em1Mvw Il2rgtm1Wjl/MvwJ |
| 028657 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav Tg(CSF1)3Sz/J |
| 014570 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(HLA-A/H2-D/B2M)1Dvs/SzJ |
| 030890 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(IL15)1Sz/SzJ |
| 035970 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(YAC128)53Hay/JnolJ |
| 035424 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Ace2em1Lutzy/J |
| 035002 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Ace2tm1(ACE2)Dwnt/J |
| 017637 | NOD.Cg-Prkdcscid Il2rgtm1Wjl H2-Ab1b-tm1Doi Tg(HLA-DRB1)31Dmz/SzJ |
| 012480 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Hprt1b-m3/EshJ |
| 034859 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CAG-cre/Esr1*)5Amc/J |
| 021937 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CAG-EGFP)1Osb/SzJ |
| 013062 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ |
| 028654 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CSF1)3Sz/SzJ |
| 029295 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(H2-Ea-HLA-DRB1*0401*)1Dv/SzJ |
| 028655 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(IL6)1Sz/SzJ |
| 027976 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(Ins2-HBEGF)6832Ugfm/SzJ |
| 017830 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(PGK1-KITLG*220)441Daw/SzJ |
| 028842 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(SERPINA1*E342K)#Slcw/SzJ |
| 005557 | NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ |
| 034569 | NOD.Cg-Prkdcscid Otcem1Bshep Il2rgtm1Wjl/J |
| 011066 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CSF2)2Ygy Tg(IL3)1Ygy Tg(KITLG)3Ygy/YgyJGckRolyJ |
| 036917 | NOD.Cg-Prltm1.1(PRL)Rui Prkdcscid Il2rgtm1Wjl/J |
| 030442 | NOD.Cg-Rag1tm1Mom Dmdmdx-4Cv Il2rgtm1Wjl/McalJ |
| 018454 | NOD.Cg-Rag1tm1Mom Fahem1Mvw Il2rgtm1Wjl/MvwJ |
| 026062 | NOD.Cg-Rag1tm1Mom Fancaem1Dvs Il2rgtm1Wjl/SzJ |
| 033127 | NOD.Cg-Rag1tm1Mom Flt3tm1Irl Mcph1Tg(HLA-A2.1)1Enge Il2rgtm1Wjl/J |
| 035855 | NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl Tg(HLA-A/H2-D/B2M)Dvs Tg(HLA-DRA,HLA-DRB1*0401)39-2Kito/J |
| 030533 | NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl Tg(SLC10A1)15Mvw/MvwJ |
| 026014 | NOD.Cg-Rag1tm1Mom KitW-41J Il2rgtm1Wjl/EavJ |
| 030443 | NOD.Cg-Rag1tm1Mom Sgcatm1Kcam Il2rgtm1Wjl/McalJ |
| 029663 | NOD.Cg-Rag1tm1Mom Dysfprmd Il2rgtm1Wjl/McalJ |
| 024099 | NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/J |
| 017914 | NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl Tg(HLA-DRA,HLA-DRB1*0401)39-2Kito/ScasJ |
| 007799 | NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ |
| 014568 | NOD.Cg-Rag1tm1Mom Ins2Akita Il2rgtm1Wjl/SzJ |
| 033704 | NOD.Cg-Tlr4lps-del Prkdcscid Il2rgtm1Wjl/SzJ |
| 028616 | NOD.Cg-Tg(CAG-FCGRT)276Dcr Fcgrttm1Dcr Prkdcscid Il2rgtm1Wjl/J |
| 026997 | NOD.Cg-Tg(CD68-EGFP)1Drg Prkdcscid Il2rgtm1Wjl/J |
| 030331 | NOD.Cg-Tg(HLA-DRA*0101,HLA-DRB1*0101)1Dmz Prkdcscid H2-Ab1b-tm1Doi Il2rgtm1Wjl/SzJ |
| 012479 | NOD.Cg-Tg(HLA-DRA*0101,HLA-DRB1*0101)1Dmz Prkdcscid Il2rgtm1Wjl/GckRolyJ |
| 034901 | NOD.Cg-Tg(K18-ACE2)2Prlmn Prkdcscid Il2rgtm1Wjl/J |
| 77 strains use this protocol | |