For in-depth product & services help, ask our
Technical Information Scientists
Stock No: 027898
Protocol 31920: Probe Assay - Generic LacZ Probe
Version 5.0
Notes
The genotyping protocol(s) presented here have been optimized for reagents and conditions used by The Jackson Laboratory (JAX).
To genotype animals, JAX recommends researchers validate the assay independently upon receipt of animals into their facility.
Reaction cycling temperature and times may require additional optimization based on the specific genotyping reagents used.
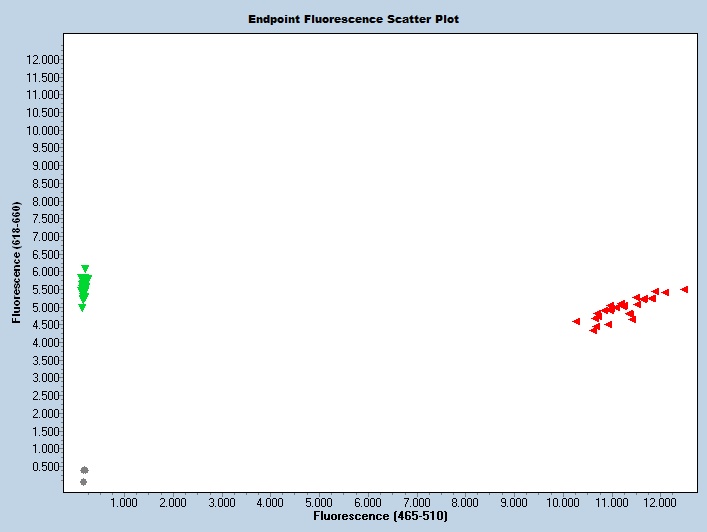
Expected Results
JAX Protocol
Protocol Primers
| Primer | 5' Label | Sequence 5' → 3' | 3' Label | Primer Type | Reaction | Note |
|---|---|---|---|---|---|---|
| 37301 | GTT TAT GCA GCA ACG AGA CG | Transgene Reverse | A | |||
| 37302 | Fluorophore-1 | CAG GAT ATG TGG CGG ATG AGC | Quencher-1 | Tg Probe | ||
| mom0095 | TGA CGG CAG TTA TCT GGA AG | Transgene Forward | A | |||
| oIMR1544 | CAC GTG GGC TCC AGC ATT | Internal Positive Control Forward | A | |||
| oIMR3580 | TCA CCA GTC ATT TCT GCC TTT G | Internal Positive Control Reverse | A | |||
| TmoIMR0105 | Fluorophore-2 | CCA ATG GTC GGG CAC TGC TCA A | Quencher-2 | IC Probe |
Reaction A
| Component | Final Concentration |
|---|---|
| Kapa Probe Fast QPCR | 1.00 X |
| ddH2O | |
| 37301 | 0.40 uM |
| mom0095 | 0.40 uM |
| oIMR1544 | 0.40 uM |
| Wt Probe | 0.15 uM |
| Mutant Probe | 0.15 uM |
| DNA |
Cycling
| Step | Temp °C | Time | Note |
|---|---|---|---|
| 1 | 95.0 | -- | |
| 2 | 95.0 | -- | |
| 3 | 60.0 | -- | |
| 4 | -- | repeat steps 2-3 for 40 cycles | |
| 5 | 40.0 | -- | Forever |
JAX uses a very high speed Taq (~1000 bp/sec), use cycling times recommended for your reagents.
Strains Using This Protocol
| Stock Number | Strain Name |
|---|---|
| 012668 | 129S.129-Lrp5tm1Grw/J |
| 038651 | 129S1.Cg-Tg(Hmgcr-lacZ)H253Sest/EmsJ |
| 002957 | B6.129-Dll1tm1Gos/J |
| 004276 | B6.129-Figntm1Frk/Frk |
| 004478 | B6.129-Foxd1tm1Lai/J |
| 003870 | B6.129-Plin1tm1Chan/J |
| 019146 | B6.129-Tmc1tm1.1Ajg/J |
| 030985 | B6.129P2(Cg)-Aph1atm1Bdes Aph1btm1Bdes Aph1ctm1Bdes/J |
| 009348 | B6.129P2(Cg)-Hprt1tm17(Ple48-lacZ)Ems/Mmjax |
| 012575 | B6.129P2(Cg)-Hprt1tm39(Ple24-lacZ)Ems/Mmjax |
| 012333 | B6.129P2(Cg)-Hprt1tm45(Ple67-lacZ)Ems/Mmjax |
| 012733 | B6.129P2(Cg)-Hprt1tm53(CAG-lacZ)Ems/Mmjax |
| 012347 | B6.129P2(Cg)-Hprt1tm64(Ple170-lacZ)Ems/Mmjax |
| 012583 | B6.129P2(Cg)-Hprt1tm68(Ple127-lacZ)Ems/Mmjax |
| 012656 | B6.129P2(Cg)-Hprt1tm70(Ple240-lacZ)Ems/Mmjax |
| 012657 | B6.129P2(Cg)-Hprt1tm71(Ple155-lacZ)Ems/Mmjax |
| 012659 | B6.129P2(Cg)-Hprt1tm73(Ple142-lacZ)Ems/Mmjax |
| 012734 | B6.129P2(Cg)-Hprt1tm74(Ple232-lacZ)Ems/Mmjax |
| 018054 | B6.129P2(Cg)-Pitx2tm1.1Dmm/J |
| 013144 | B6.129P2-Aim2Gt(CSG445)Byg/J |
| 005801 | B6.129P2-Esrratm1Dgen/J |
| 005814 | B6.129P2-Grm1tm1Dgen/J |
| 034597 | B6.129P2-Kif13aGt(AF0553)Wtsi/AhcJ |
| 030421 | B6.129P2-Mbd2tm1Bh/JoezJ |
| 023209 | B6.129S-Sfrp1tm1Jrbn/PcmaJ |
| 004950 | B6.129S1-Syt7tm1Nan/J |
| 036325 | B6.129S6-Mecomtm1Aspe/Mmjax |
| 005876 | B6.CAST-(rs4222996-rs13476494)/LusJ |
| 019547 | B6.Cg-Cacna1ctm2Itl/J |
| 017762 | B6.Cg-Cacna1ftm1.2Sdie/J |
| 030065 | B6.Cg-Serpinf1tm1Craw/J |
| 033441 | B6.Cg-Slc15a1tm1Dgen Tg(SLC15A1)1Dsmi/Mmjax |
| 006477 | B6.Cg-Tg(CAG-lacZ-WGA)330Bbm/J |
| 033345 | B6.Cg-Tg(Cdh5-COP4*L132C,-lacZ)B28-2Mik/J |
| 032889 | B6.Cg-Tg(Cdh5-RHO/Adrb2,-lacZ)B27-3Mik/J |
| 006229 | B6.Cg-Tg(DRE-lacZ)2Gswz/J |
| 033344 | B6.Cg-Tg(Hcn4-COP4*L132C,-lacZ)B17-2Mik/J |
| 024050 | B6.Cg-Tg(Hmgcr-lacZ)H253Sest/J |
| 023757 | B6.Cg-Tg(tetO-tetX,lacZ)1Gogo/UmriJ |
| 028353 | B6.Cg-Tg(Vil1-EGFP)106Gum/J |
| 018625 | B6.FVB-Tg(Fabp4-lacZ)4Mosh/J |
| 025740 | B6;129-Prg4tm2Mawa/J |
| 008704 | B6;129-Smn1tm6(SMN2)Mrph/J |
| 006251 | B6;129-Tor1atm1Wtd/J |
| 025197 | B6;129-Vmn1r51tm1Dlc/J |
| 025196 | B6;129-Vmn1r51tm2.1Dlc/J |
| 017524 | B6;129-Tg(CMV-Bgeo,-WGA,-ALPP)1Mgmj/J |
| 038621 | B6;129P2-Ctcftm2.2Gal/J |
| 024288 | B6;129P2-Faptm2Schn/J |
| 006744 | B6;129P2-Gt(ROSA)26Sortm1Mom/MomJ |
| 032534 | B6;129P2-Maltm1Nsw/J |
| 006665 | B6;129P2-Or8a1tm13(Olr226)Mom/MomJ |
| 012850 | B6;129P2-TardbpGt(RRB030)Byg/J |
| 026012 | B6;129P2-Uts2rtm1Djbe/J |
| 006465 | B6;CBA-Tg(CAG-lacZ-WGA)330Bbm/J |
| 007972 | B6;CBA-Tg(Olfr151-taulacZ)4Mom/MomJ |
| 006680 | B6;CBA-Tg(Olfr16*,taulacZ)19Mom/MomJ |
| 006672 | B6;CBA-Tg(Olfr16*,taulacZ)7Mom/MomJ |
| 007973 | B6;CBA-Tg(Olfr16-taulacZ)1Mom/MomJ |
| 007974 | B6;CBA-Tg(Olfr160-taulacZ)V4-7Mom/MomJ |
| 007976 | B6;CBA-Tg(Olfr713-taulacZ)4Mom/MomJ |
| 007975 | B6;CBA-Tg(OR8A1-taulacZ)1Mom/MomJ |
| 004141 | B6;CBA-Tg(UAS-lacZ)65Rth/J |
| 008344 | B6;DBA-Tg(Fos-tTA,Fos-EGFP*)1Mmay Tg(tetO-lacZ,tTA*)1Mmay/J |
| 018627 | B6;SJL-Tg(Myl1-lacZ)1Ibdml/J |
| 002621 | B6;SJL-Tg(tetop-lacZ)2Mam/J |
| 003299 | B6;SWJ-Tg(TIMP3-lacZ)7Jeb/J |
| 037014 | B6J.129P2-Pofut2Gt(RST434)Byg/BchMmjax |
| 024866 | B6J.B6N(Cg)-Sec24ctm1a(EUCOMM)Wtsi/J |
| 034731 | B6N(129S4)-Srd5a3tm1c(EUCOMM)Wtsi/J |
| 030130 | B6N.129-Dkk2tm1Dwu/J |
| 030043 | B6N;129-LrbaGt(OST69142)Lex/WgkJ |
| 002754 | C57BL/6-Tg(LacZpl)60Vij/J |
| 023048 | C57BL/6-Tg(Peg3-lacZ)26Sass/J |
| 027898 | C57BL/6J-Tg(CAG-lacZ,-FUS,-EGFP)629Gyu/J |
| 037062 | FVB.129S-Neu1tm1Adz/J |
| 005025 | FVB.Cg-Grm7Tg(SMN2)89Ahmb Smn1tm1Msd Tg(SMN2*delta7)4299Ahmb/J |
| 031323 | FVB/N-Tg(Bmp5,Hspa1b-lacZ)639Kng/J |
| 031325 | FVB/N-Tg(Bmp5-Hmgcll1,Hspa1b-lacZ)1Kng/J |
| 031324 | FVB/N-Tg(Bmp5-Hmgcll1,Hspa1b-lacZ)665Kng/J |
| 026047 | FVB/N-Tg(Isl1-lacZ)5Cka/Mmjax |
| 002856 | FVB/N-Tg(TIE2-lacZ)182Sato/J |
| 035185 | STOCK Adgrl1tm2c(EUCOMM)Hmgu/SudJ |
| 026229 | STOCK Akap12tm1Ihg Rb1tm2Brn Tg(Pbsn-cre)4Prb/J |
| 035392 | STOCK Gsctm2Bhr/J |
| 024284 | STOCK Hcn4tm3Sev/J |
| 013764 | STOCK Hprt1tm57(Ple26-lacZ)Ems/Mmjax |
| 012354 | STOCK Hprt1tm66(Ple5-lacZ)Ems/Mmjax |
| 012584 | STOCK Hprt1tm69(Ple134-lacZ)Ems/Mmjax |
| 013087 | STOCK Mir7-2tm1Mtm/Mmjax |
| 006578 | STOCK Myoz2tm1Eno/J |
| 006686 | STOCK Or6b9tm1Mom/MomJ |
| 006613 | STOCK Tg(CAG-Bgeo,-Tle1,-ALPP)1Lbe/J |
| 004623 | STOCK Tg(TCF/Lef1-lacZ)34Efu/J |
| 005728 | STOCK Tg(tetO-Ipf1,lacZ)958.1Macd/J |
| 95 strains use this protocol | |