For in-depth product & services help, ask our
Technical Information Scientists
Stock No: 023099
Protocol 18501: QPCR Assay - c9ORF72-qPCR
Version 1.0
Notes
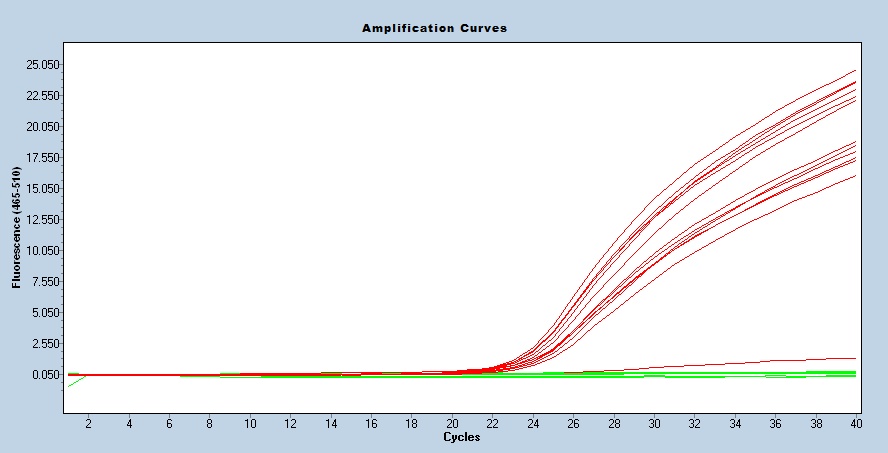
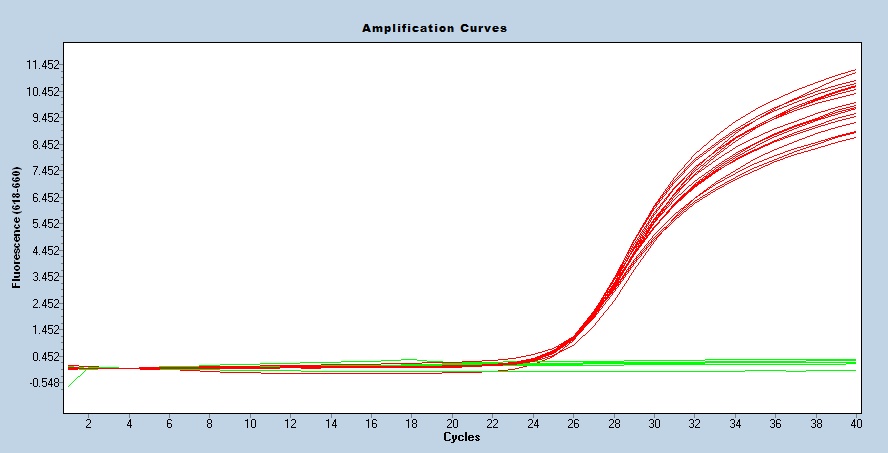
Taqman qPCR protocols are run on a real time PCR instrument. Use an appropriate instrument specific Fluorophore/Quencher combination. The transgene genotype is determined by comparing ΔCt values of each unknown sample against known homozygous and hemizygous controls, using appropriate endogenous references.
Genotyping by PCR is performed to determine the absence or presence of the transgene. This strain has some variability in repeat size, therefore Southern blotting is performed to accurately determine the size, in kilobases, of the repeat within the transgene.
The genotyping protocol(s) presented here have been optimized for reagents and conditions used by The Jackson Laboratory (JAX).
To genotype animals, JAX recommends researchers validate the assay independently upon receipt of animals into their facility.
Reaction cycling temperature and times may require additional optimization based on the specific genotyping reagents used.
Expected Results
Tg= 150 bp
IPC = 74 bp
JAX Protocol
Protocol Primers
| Primer | 5' Label | Sequence 5' → 3' | 3' Label | Primer Type | Reaction | Note |
|---|---|---|---|---|---|---|
| 27157 | AGG AGC TGG TGA GTT TCA AC | Transgene Forward | A | |||
| 27158 | GCT CCC ACA GTG TAA TTC TCT | Transgene Reverse | A | |||
| 27159 | Fluorophore-1 | ACT GCC CGT TGT GAC CTG AGA C | Quencher-1 | |||
| oIMR1544 | CAC GTG GGC TCC AGC ATT | Internal Positive Control Forward | A | |||
| oIMR3580 | TCA CCA GTC ATT TCT GCC TTT G | Internal Positive Control Reverse | A | |||
| TmoIMR0105 | Fluorophore-2 | CCA ATG GTC GGG CAC TGC TCA A | Quencher-2 |
Reaction A
| Component | Final Concentration |
|---|---|
| ddH2O | |
| Kapa Probe Fast QPCR | 1.00 X |
| 27157 | 0.40 uM |
| 27158 | 0.40 uM |
| oIMR1544 | 0.40 uM |
| oIMR3580 | 0.40 uM |
| Tg Probe | 0.15 uM |
| IC Probe | 0.15 uM |
| DNA |
Cycling
| Step | Temp °C | Time | Note |
|---|---|---|---|
| 1 | 95.0 | -- | |
| 2 | 95.0 | -- | |
| 3 | 60.0 | -- | repeat steps 2-3 for 40 cycles |
JAX uses a very high speed Taq (~1000 bp/sec), use cycling times recommended for your reagents.
Strains Using This Protocol
This is the only strain that uses this protocol.