For in-depth product & services help, ask our
Technical Information Scientists
Stock No: 008870
Protocol 29081: Standard PCR Assay - Generic Cre Melt Curve Analysis
Version 2.3
Notes
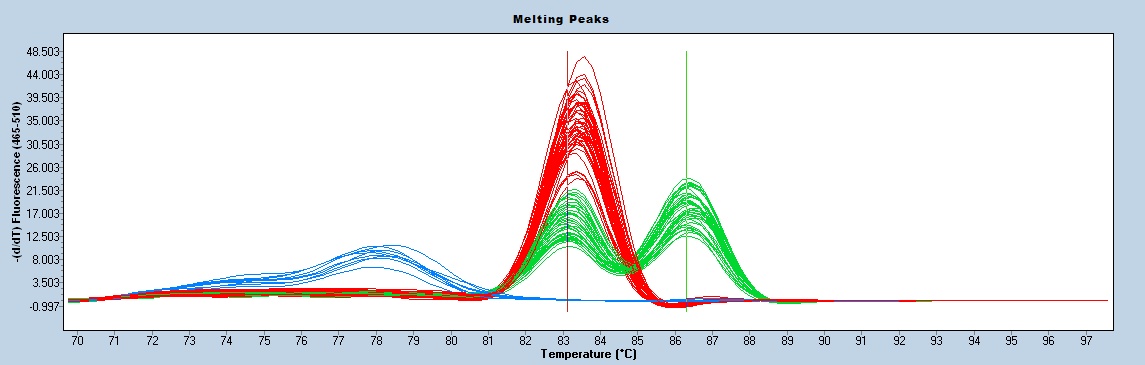
Melting curve analysis is done using a Roche Light Cycler 480.
This assay will NOT distinguish hemizygous from homozygous transgenic animals.
This assay will NOT distinguish hemizygous from homozygous transgenic animals.
The genotyping protocol(s) presented here have been optimized for reagents and conditions used by The Jackson Laboratory (JAX).
To genotype animals, JAX recommends researchers validate the assay independently upon receipt of animals into their facility.
Reaction cycling temperature and times may require additional optimization based on the specific genotyping reagents used.
Expected Results
Transgene Tm = 83°C +/- 1.0°C
Internal positive control Tm = 86.5°C+/- 1.0°C
Transgene = ~100 bp
Internal positive control = 324 bp
JAX Protocol
Protocol Primers
| Primer | 5' Label | Sequence 5' → 3' | 3' Label | Primer Type | Reaction | Note |
|---|---|---|---|---|---|---|
| oIMR1084 | GCG GTC TGG CAG TAA AAA CTA TC | Transgene Forward | A | Cre F | ||
| oIMR1085 | GTG AAA CAG CAT TGC TGT CAC TT | Transgene Reverse | A | Cre R | ||
| oIMR7338 | CTA GGC CAC AGA ATT GAA AGA TCT | Internal Positive Control Forward | A | |||
| oIMR7339 | GTA GGT GGA AAT TCT AGC ATC ATC C | Internal Positive Control Reverse | A |
Reaction A
| Component | Final Concentration |
|---|---|
| ddH2O | |
| Kapa 2G HS buffer | 1.30 X |
| MgCl2 | 2.60 mM |
| dNTP KAPA | 0.26 mM |
| oIMR1084 | 0.50 uM |
| oIMR1085 | 0.50 uM |
| oIMR7338 | 0.50 uM |
| oIMR7339 | 0.50 uM |
| Glycerol | 6.50 % |
| Dye | 1.00 X |
| Kapa 2G HS taq polym | 0.03 U/ul |
| DNA |
Cycling
| Step | Temp °C | Time | Note |
|---|---|---|---|
| 1 | 94.0 | -- | |
| 2 | 94.0 | -- | |
| 3 | 65.0 | -- | -0.5 C per cycle decrease |
| 4 | 68.0 | -- | |
| 5 | -- | repeat steps 2-4 for 10 cycles (Touchdown) | |
| 6 | 94.0 | -- | |
| 7 | 60.0 | -- | |
| 8 | 72.0 | -- | |
| 9 | -- | repeat steps 6-8 for 28 cycles | |
| 10 | 72.0 | -- | |
| 11 | 10.0 | -- | hold |
JAX uses a very high speed Taq (~1000 bp/sec), use cycling times recommended for your reagents.
JAX uses a 'touchdown' cycling protocol and therefore has not calculated the optimal annealing temperature for each set of primers.
Strains Using This Protocol
| Stock Number | Strain Name |
|---|---|
| 005989 | 129;FVB-Tg(PTH-cre)4167Slib/J |
| 017789 | 129S-Slc11a2tm2Nca/J |
| 007179 | 129S.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/J |
| 007915 | 129S.FVB-Tg(Amh-cre)8815Reb/J |
| 004302 | 129S1/Sv-Hprt1tm1(CAG-cre)Mnn/J |
| 008523 | 129S6.Cg-Tg(NPHS2-cre)295Lbh/BroJ |
| 032051 | B6(129S4)-Kittm1.1(cre)Jmol/J |
| 037511 | B6(129S4)-Pou2f3tm1.1(cre/ERT2)Imt/J |
| 024976 | B6(129S4)-Tspotm1.1Maf/J |
| 009575 | B6(129S4)-Et(cre/ERT2)119Rdav/J |
| 009580 | B6(129S4)-Et(cre/ERT2)1382Rdav/J |
| 009581 | B6(129S4)-Et(cre/ERT2)1642Rdav/J |
| 009582 | B6(129S4)-Et(cre/ERT2)1645Rdav/J |
| 009583 | B6(129S4)-Et(cre/ERT2)1957Rdav/J |
| 009584 | B6(129S4)-Et(cre/ERT2)2007Rdav/J |
| 009585 | B6(129S4)-Et(cre/ERT2)2047Rdav/J |
| 009574 | B6(129S4)-Et(cre/ERT2)21Rdav/J |
| 009577 | B6(129S4)-Et(cre/ERT2)296Rdav/J |
| 009578 | B6(129S4)-Et(cre/ERT2)398Rdav/J |
| 009573 | B6(129S4)-Et(cre/ERT2)4Rdav/J |
| 009579 | B6(129S4)-Et(cre/ERT2)837Rdav/J |
| 038514 | B6(129S6)-Tg(Slc6a5-cre/ERT2)#Uze/J |
| 026512 | B6(Cg)-Tg(TPO-cre/ERT2)1139Tyj/J |
| 021012 | B6(D2)-Tg(CAG-Brainbow1.0)3Eggn/J |
| 037052 | B6(FVB)-Tg(Sprr2f-cre)2Dcas/J |
| 018057 | B6.129(Cg)-Cul7tm2.1Jdec/J |
| 008320 | B6.129(Cg)-Leprtm2(cre)Rck/J |
| 033964 | B6.129(Cg)-Ptf1atm2(cre/ESR1)Cvw Krastm4Tyj Ptentm1Hwu/J |
| 036564 | B6.129(Cg)-Scn10atm2(cre)Jwo/TjpJ |
| 008463 | B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J |
| 018445 | B6.129-Mre11atm1.1Rchd/J |
| 025567 | B6.129-Olig2tm1.1(cre)Wdr/J |
| 005697 | B6.129-Otx1tm4(cre)Asim/J |
| 004146 | B6.129-Tg(Pcp2-cre)2Mpin/J |
| 021160 | B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ |
| 018054 | B6.129P2(Cg)-Pitx2tm1.1Dmm/J |
| 023726 | B6.129P2-C1qatm1.1Tenn/J |
| 016934 | B6.129P2-Lgr6tm2.1(cre/ERT2)Cle/J |
| 018997 | B6.129S(FVB)-Lhx3tm1.1Sjr/J |
| 016132 | B6.129S4-Perptm2Att/J |
| 006447 | B6.129S6(CBA)-Cebpatm1Dgt/J |
| 012900 | B6.129S6-Stat5btm1Mam Stat5atm2Mam/Mmjax |
| 006054 | B6.C-Tg(CMV-cre)1Cgn/J |
| 008601 | B6.Cg-7630403G23RikTg(Th-cre)1Tmd/J |
| 006230 | B6.Cg-Cebpatm1Dgt Tg(Mx1-cre)1Cgn/J |
| 036450 | B6.Cg-Foxl1tm2(cre/ERT2)Khk/Mmjax |
| 009107 | B6.Cg-H2az2Tg(Wnt1-cre)11Rth Tg(Wnt1-GAL4)11Rth/J |
| 032923 | B6.Cg-Kittm1(cre)Htng/J |
| 028536 | B6.Cg-Ndnftm1.1(folA/cre)Hze/J |
| 030544 | B6.Cg-Npy1rtm1.1(cre/GFP)Rpa/J |
| 031485 | B6.Cg-Pou4f2tm1(cre)Bnt/SjJ |
| 017889 | B6.Cg-Shank3tm1.1Bux/J |
| 022865 | B6.Cg-Trib2tm1.1(cre/ERT2)Hze/J |
| 017346 | B6.Cg-Tg(A930038C07Rik-cre)1Aibs/J |
| 006149 | B6.Cg-Tg(ACTA1-cre)79Jme/J |
| 030215 | B6.Cg-Tg(Amigo2-cre)1Sieg/J |
| 009352 | B6.Cg-Tg(CDX2-cre*)189Erf/J |
| 006368 | B6.Cg-Tg(Cr2-cre)3Cgn/J |
| 029594 | B6.Cg-Tg(Dmp1-cre/ERT2)D77Pdp/J |
| 006663 | B6.Cg-Tg(Eno2-cre)39Jme/J |
| 035668 | B6.Cg-Tg(Gata1-cre)1Sho/MdfJ |
| 009642 | B6.Cg-Tg(GH1-cre)1Sac/J |
| 003573 | B6.Cg-Tg(Ins2-cre)25Mgn/J |
| 005584 | B6.Cg-Tg(Prrx1-cre)1Cjt/J |
| 003967 | B6.Cg-Tg(Rbp3-cre)528Jxm/J |
| 021614 | B6.Cg-Tg(S100A8-cre,-EGFP)1Ilw/J |
| 004128 | B6.Cg-Tg(Tek-cre)12Flv/J |
| 008863 | B6.Cg-Tg(Tek-cre)1Ywa/J |
| 012328 | B6.Cg-Tg(Tyr-cre/ERT2)13Bos/J |
| 035670 | B6.Cg-Tg(VAV1-cre)1Graf/MdfJ |
| 020282 | B6.Cg-Tg(Vil1-cre/ERT2)23Syr/J |
| 006451 | B6.FVB(129X1)-Tg(Sim1-cre)1Lowl/J |
| 006137 | B6.FVB-Tg(Cdh5-cre)7Mlia/J |
| 018980 | B6.FVB-Tg(Ddx4-cre)1Dcas/KnwJ |
| 003724 | B6.FVB-Tg(EIIa-cre)C5379Lmgd/J |
| 011069 | B6.FVB-Tg(Gh1-cre)bKnmn/J |
| 011038 | B6.FVB-Tg(Myh6-cre)2182Mds/J |
| 010714 | B6.FVB-Tg(Pomc-cre)1Lowl/J |
| 003394 | B6.FVB-Tg(Zp3-cre)3Mrt/J |
| 017601 | B6;129-Eif2s1tm1Rjk/J |
| 016210 | B6;129-Lrrk2tm3.1Shn/J |
| 018319 | B6;129-Plxnd1tm1.1Tmj/J |
| 009600 | B6;129-Six2tm3(EGFP/cre/ERT2)Amc/J |
| 018773 | B6;129-Sult2b1tm1Rus/J |
| 022864 | B6;129S-Rasgrf2tm1(cre/folA)Hze/J |
| 017593 | B6;129S-Sox2tm1(cre/ERT2)Hoch/J |
| 009388 | B6;129S1-Osr2tm2(cre)Jian/J |
| 036300 | B6;129S4-Cux1tm1(cre/ERT2)Zjh/J |
| 018019 | B6;129S4-Srsf2tm1Xdfu/J |
| 009576 | B6;129S4-Et(cre/ERT2)278Rdav/J |
| 017495 | B6;129S7-Crim1tm1(GFP/cre/ERT2)Pzg/J |
| 018937 | B6;C-Tg(Erv4-cre/ERT2*)657Slp/J |
| 009616 | B6;C3-Tg(A930038C07Rik-cre)4Aibs/J |
| 008844 | B6;C3-Tg(Ctgf-cre)2Aibs/J |
| 008839 | B6;C3-Tg(Cyp39a1-cre)1Aibs/J |
| 009117 | B6;C3-Tg(Cyp39a1-cre)7Aibs/J |
| 009111 | B6;C3-Tg(Scnn1a-cre)1Aibs/J |
| 009112 | B6;C3-Tg(Scnn1a-cre)2Aibs/J |
| 009613 | B6;C3-Tg(Scnn1a-cre)3Aibs/J |
| 009103 | B6;C3-Tg(Wfs1-cre/ERT2)3Aibs/J |
| 025809 | B6;CBA-Tg(Fgfr3-icre/ERT2)4-2Wdr/J |
| 024926 | B6;D2-Tg(Fshr-cre)1Ldu/J |
| 003466 | B6;D2-Tg(Sycp1-cre)4Min/J |
| 015855 | B6;DBA-Tg(Upk3a-GFP/cre/ERT2)26Amc/J |
| 018402 | B6;FVB-Nrn1tm1.2Ndiv/J |
| 003734 | B6;FVB-Tg(GZMB-cre)1Jcb/J |
| 004426 | B6;SJL-Tg(Cga-cre)3Sac/J |
| 003554 | B6;SJL-Tg(Col2a1-cre)1Bhr/J |
| 017738 | B6;SJL-Tg(Foxl1-cre)1Khk/J |
| 005249 | B6;SJL-Tg(Krt1-15-cre/PGR*)22Cot/J |
| 003552 | B6129-Tg(Wap-cre)11738Mam/J |
| 007252 | B6Ei.129S4-Tg(Prm-cre)58Og/EiJ |
| 017741 | B6N.129(Cg)-Mecp2tm1.1Joez/J |
| 017704 | B6N.129-Pik3catm1Jjz/J |
| 016881 | B6N.Cg-Adgra2tm1.1Bstc/J |
| 019102 | B6N.Cg-Tg(CAG-cre/Esr1*)5Amc/CjDswJ |
| 018966 | B6N.Cg-Tg(Camk2a-cre)T29-1Stl/J |
| 018965 | B6N.Cg-Tg(Fabp4-cre)1Rev/J |
| 018960 | B6N.Cg-Tg(Ins2-cre)25Mgn/J |
| 018964 | B6N.Cg-Tg(KRT14-cre)1Amc/J |
| 019103 | B6N.Cg-Tg(Nes-cre)1Kln/CjDswJ |
| 003465 | BALB/c-Tg(CMV-cre)1Cgn/J |
| 013756 | C.129S-Batf3tm1Kmm/J |
| 004126 | C.Cg-Cd19tm1(cre)Cgn Ighb/J |
| 005673 | C.Cg-Tg(Mx1-cre)1Cgn/J |
| 006244 | C.Cg-Tg(tetO-cre)1Jaw/J |
| 009155 | C57BL/6-Cldn6tm1(cre)Dkwu/J |
| 035489 | C57BL/6-Elmo1tm1Mki/J |
| 038053 | C57BL/6-Oprm1tm2.1(EGFP/cre)Ics/KffJ |
| 016097 | C57BL/6-Tg(Car1-cre)5Flt/J |
| 008870 | C57BL/6-Tg(Hspa2-cre)1Eddy/J |
| 024034 | C57BL/6-Tg(Pmch-cre)1Rck/J |
| 036429 | C57BL/6J-Gnetm1Dar/J |
| 032080 | C57BL/6J-Tg(Cd8a*-cre)B8Asin/J |
| 018899 | C57BL/6J-Tg(Krt6,-cre,-Cerulean)5Grsr/Grsr |
| 021582 | C57BL/6J-Tg(Mchr1-cre)1Emf/J |
| 022883 | C57BL/6J-Tg(Six6-cre)3Grsr/GrsrJ |
| 027789 | C57BL/6N-Fgd5tm3(cre/ERT2)Djr/J |
| 023453 | C57BL/6N-Lhx8tm1(cre)Grsr/2GrsrJ |
| 012686 | C57BL/6N-Tg(Ppp1r2-cre)4127Nkza/J |
| 012929 | FVB(Cg)-Tg(Dhh-cre)1Mejr/J |
| 004600 | FVB-Tg(GFAP-cre)25Mes/J |
| 005025 | FVB.Cg-Grm7Tg(SMN2)89Ahmb Smn1tm1Msd Tg(SMN2*delta7)4299Ahmb/J |
| 006139 | FVB.Cg-Tg(ACTA1-cre)79Jme/J |
| 006297 | FVB.Cg-Tg(Eno2-cre)39Jme/J |
| 018394 | FVB.Cg-Tg(KRT5-cre/ERT2)2Ipc/JeldJ |
| 008244 | FVB.Cg-Tg(tetO-cre)1Jaw/J |
| 003314 | FVB/N-Tg(EIIa-cre)C5379Lmgd/J |
| 003377 | FVB/N-Tg(Zp3-cre)3Mrt/J |
| 017678 | FVB;129-Pink1tm1Aub Tg(Prnp-SNCA*A53T)AAub/J |
| 023325 | FVB;B6-Tg(Pbsn-cre)20Fwan/J |
| 034859 | NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CAG-cre/Esr1*)5Amc/J |
| 034795 | NOD.Cg-Tg(CAG-cre/Esr1*)5Amc/J |
| 013251 | NOD.FVB-Tg(EIIa-cre)C5379Lmgd/J |
| 004986 | NOD/ShiLt-Tg(Ins2-cre)3Lt/LtJ |
| 003855 | NOD/ShiLt-Tg(Ins2-cre)5Lt/LtJ |
| 035185 | STOCK Adgrl1tm2c(EUCOMM)Hmgu/SudJ |
| 036942 | STOCK Agertm2.1(cre/ERT2)Blh/2J |
| 037028 | STOCK Calcrtm1.1(cre)Mgmj/J |
| 007916 | STOCK En1tm2(cre)Wrst/J |
| 007913 | STOCK Gli1tm3(cre/ERT2)Alj/J |
| 008783 | STOCK Grm7Tg(SMN2)89Ahmb Smn1tm3(SMN2/Smn1)Mrph Tg(CAG-cre/Esr1*)5Amc Tg(SMN2*delta7)4299Ahmb/J |
| 022793 | STOCK Gt(ROSA)26Sortm1(LRRK2*R1441C)Djmo/J |
| 003829 | STOCK H2az2Tg(Wnt1-cre)11Rth Tg(Wnt1-GAL4)11Rth/J |
| 017298 | STOCK Hmgn5tm1.1Mbus/J |
| 035978 | STOCK Hoxb8tm2.1(cre)Mrc/J |
| 032429 | STOCK Krastm4Tyj Trp53tm1Brn Tg(Pdx1-cre/Esr1*)#Dam/J |
| 018418 | STOCK Lrig1tm1.1(cre/ERT2)Rjc/J |
| 016945 | STOCK Mir138-1tm1Mtm/Mmjax |
| 004192 | STOCK Mttptm2Sgy Ldlrtm1Her Apobtm2Sgy Tg(Mx1-cre)1Cgn/J |
| 037139 | STOCK Myl1tm1(cre)Sjb Atg7tm1Tchi Gaatm1Rabn/PuroMmjax |
| 035303 | STOCK Obscntm1Chen/J |
| 035925 | STOCK Opn4tm1(cre)Saha/J |
| 035045 | STOCK Oprk1tm1.1(cre)Sros/J |
| 030543 | STOCK Oxtrtm1(cre/GFP)Rpa/J |
| 022972 | STOCK Pros1tm1Grl/J |
| 019378 | STOCK Ptf1atm2(cre/ESR1)Cvw/J |
| 013086 | STOCK Rxratm1Krc/J |
| 020080 | STOCK Slc6a3tm1(cre)Xz/J |
| 023405 | STOCK Tfap2atm1Hsv/J |
| 036906 | STOCK Tmem178btm1c(KOMP)Wtsi Tmem178tm1.1Sud/J |
| 010911 | STOCK Wt1tm1(EGFP/cre)Wtp/J |
| 007684 | STOCK Tg(Atoh1-cre/Esr1*)14Fsh/J |
| 004453 | STOCK Tg(CAG-cre/Esr1*)5Amc/J |
| 035566 | STOCK Tg(CAG-mRuby2*,-mClover*)1Srgt/J |
| 009615 | STOCK Tg(Cartpt-cre)1Aibs/J |
| 017336 | STOCK Tg(Cd4-cre)1Cwi/BfluJ |
| 005105 | STOCK Tg(Chx10-EGFP/cre,-ALPP)2Clc/J |
| 008852 | STOCK Tg(En2-cre)22Alj/J |
| 005938 | STOCK Tg(Eno2-cre)39Jme/J |
| 022763 | STOCK Tg(Eno2-cre/ERT2)1Pohlk/J |
| 027681 | STOCK Tg(Hnf1b-cre/ERT2)1Jfer/J |
| 019380 | STOCK Tg(IVL-cre/ERT2)1Blpn/J |
| 008582 | STOCK Tg(Kcnc2-Cre)K128Stl/LetJ |
| 005107 | STOCK Tg(KRT14-cre/ERT)20Efu/J |
| 017948 | STOCK Tg(KRT18-cre/ERT2)23Blpn/J |
| 009643 | STOCK Tg(Lhb-cre)1Sac/J |
| 023724 | STOCK Tg(mI56i-cre,EGFP)1Kc/J |
| 003553 | STOCK Tg(MMTV-cre)4Mam/J |
| 008847 | STOCK Tg(Msx2-cre/ERT)27Alj/J |
| 009074 | STOCK Tg(Myh6-cre)1Jmk/J |
| 009102 | STOCK Tg(Nefh-cre)12Kul/J |
| 028364 | STOCK Tg(Neurod1-cre)1Able/J |
| 005667 | STOCK Tg(Neurog3-cre)C1Able/J |
| 006207 | STOCK Tg(Pcp2-cre)1Amc/J |
| 009606 | STOCK Tg(Six2-EGFP/cre)1Amc/J |
| 032646 | STOCK Tg(Spo11-cre)1Rsw/PecoJ |
| 006224 | STOCK Tg(tetO-cre)1Jaw/J |
| 024240 | STOCK Tg(Tnnt2-cre)5Blh/JiaoJ |
| 008851 | STOCK Tg(Wnt1-cre/ERT)1Alj/J |
| 018281 | STOCK Tg(Wnt7a-EGFP/cre)#Bhr/Mmjax |
| 211 strains use this protocol | |