For in-depth product & services help, ask our
Technical Information Scientists
Stock No: 002664
Protocol 25732: Standard PCR Assay - Cd8a<tm1Mak>
Version 5.2
Notes
The genotyping protocol(s) presented here have been optimized for reagents and conditions used by The Jackson Laboratory (JAX).
To genotype animals, JAX recommends researchers validate the assay independently upon receipt of animals into their facility.
Reaction cycling temperature and times may require additional optimization based on the specific genotyping reagents used.
Expected Results
Het = middle melt curves
Hom= upper melt curves
WT = lowest melt curves
Hom= upper melt curves
WT = lowest melt curves
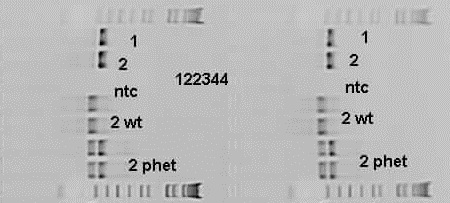
HET = 265 bp and 343 bp
MUT = 343 bp
WT = 265 bp
MUT = 343 bp
WT = 265 bp
JAX Protocol
Protocol Primers
| Primer | 5' Label | Sequence 5' → 3' | 3' Label | Primer Type | Reaction | Note |
|---|---|---|---|---|---|---|
| oIMR1098 | GAC CTG GTA TGT GAA GTG TTG G | Common | A | |||
| oIMR1099 | ACA TCA CCG AGT TGC TGA TG | Wild type | A | |||
| oIMR6828 | CAT AGC GTT GGC TAC CCG TG | Mutant | A | Neo F |
Reaction A
| Component | Final Concentration |
|---|---|
| ddH2O | |
| Kapa 2G HS buffer | 1.30 X |
| MgCl2 | 2.60 mM |
| dNTP KAPA | 0.26 mM |
| oIMR1098 | 0.50 uM |
| oIMR1099 | 0.50 uM |
| oIMR6828 | 0.50 uM |
| Glycerol | 6.50 % |
| Dye | 1.00 X |
| Kapa 2G HS taq polym | 0.03 U/ul |
| DNA |
Cycling
| Step | Temp °C | Time | Note |
|---|---|---|---|
| 1 | 94.0 | -- | |
| 2 | 94.0 | -- | |
| 3 | 65.0 | -- | -0.5 C per cycle decrease |
| 4 | 68.0 | -- | |
| 5 | -- | repeat steps 2-4 for 10 cycles (Touchdown) | |
| 6 | 94.0 | -- | |
| 7 | 60.0 | -- | |
| 8 | 72.0 | -- | |
| 9 | -- | repeat steps 6-8 for 28 cycles | |
| 10 | 72.0 | -- | |
| 11 | 10.0 | -- | hold |
JAX uses a very high speed Taq (~1000 bp/sec), use cycling times recommended for your reagents.
JAX uses a 'touchdown' cycling protocol and therefore has not calculated the optimal annealing temperature for each set of primers.